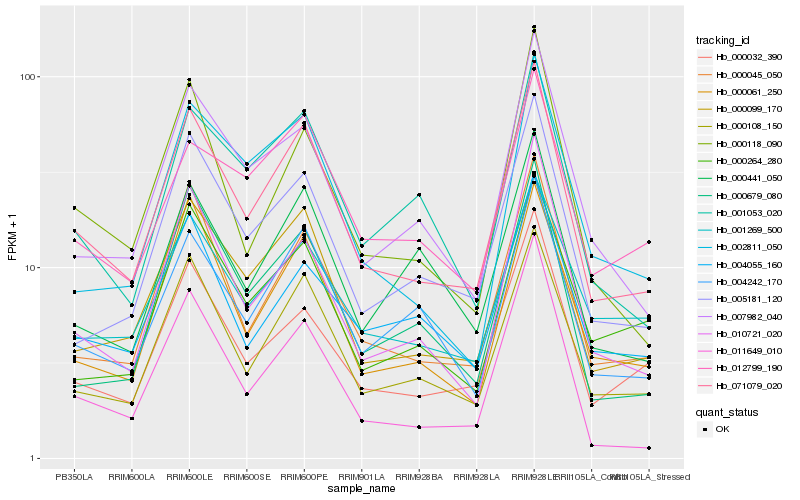
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071079_020 |
0.0 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 2 |
Hb_000108_150 |
0.073683818 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 3 |
Hb_000045_050 |
0.0889933615 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000099_170 |
0.0922410808 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 5 |
Hb_000061_250 |
0.0996896494 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 6 |
Hb_004242_170 |
0.0999489287 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 7 |
Hb_005181_120 |
0.1018363198 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_010721_020 |
0.1075789438 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007982_040 |
0.1087388892 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000032_390 |
0.1096590083 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 11 |
Hb_000118_090 |
0.1120626328 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001053_020 |
0.1143091651 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 13 |
Hb_000679_080 |
0.1172779936 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011649_010 |
0.1193414791 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 15 |
Hb_002811_050 |
0.1204517232 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 16 |
Hb_001269_500 |
0.1212208109 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 17 |
Hb_004055_160 |
0.1215528461 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_012799_190 |
0.1219556298 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 19 |
Hb_000264_280 |
0.122798102 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 20 |
Hb_000441_050 |
0.1242060398 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |