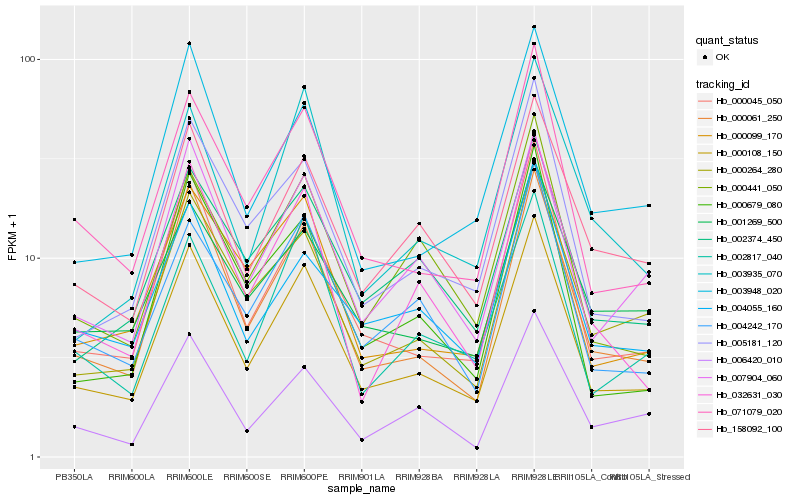
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_150 |
0.0 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 2 |
Hb_000061_250 |
0.0595748299 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 3 |
Hb_000045_050 |
0.0699646547 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_071079_020 |
0.073683818 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 5 |
Hb_005181_120 |
0.0860196145 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000264_280 |
0.0899994969 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 7 |
Hb_007904_060 |
0.0906879337 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 8 |
Hb_004242_170 |
0.0907634169 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 9 |
Hb_000099_170 |
0.0939306824 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 10 |
Hb_032631_030 |
0.0955465543 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 11 |
Hb_158092_100 |
0.0958129006 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 12 |
Hb_003948_020 |
0.0963950621 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 13 |
Hb_002817_040 |
0.0999454814 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 14 |
Hb_002374_450 |
0.1026284107 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000679_080 |
0.1027708374 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000441_050 |
0.1031784657 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 17 |
Hb_004055_160 |
0.1057767572 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006420_010 |
0.1087088807 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 19 |
Hb_001269_500 |
0.1109313447 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 20 |
Hb_003935_070 |
0.1112781849 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |