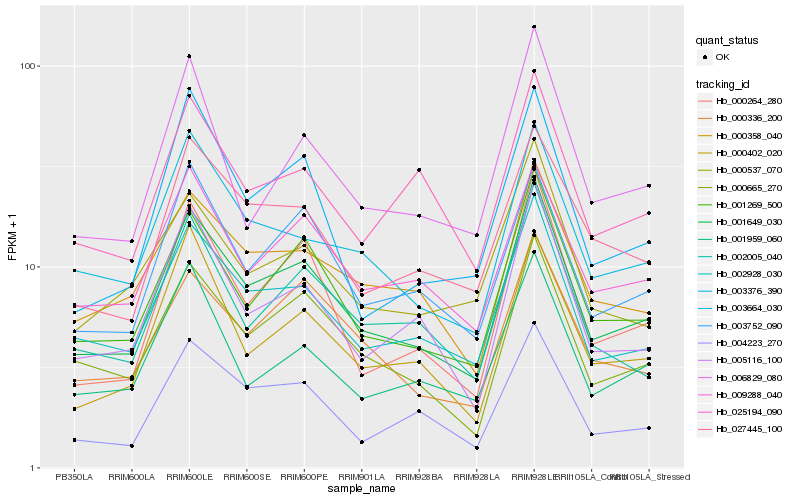
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001649_030 |
0.0 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001269_500 |
0.0657737463 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 3 |
Hb_002928_030 |
0.0683439991 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 4 |
Hb_000665_270 |
0.0780677465 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 5 |
Hb_003752_090 |
0.0897321242 |
- |
- |
chitinase, putative [Ricinus communis] |
| 6 |
Hb_000358_040 |
0.089912513 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003376_390 |
0.0904981957 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005116_100 |
0.0910270173 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 9 |
Hb_000336_200 |
0.0930828129 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_000264_280 |
0.0933973741 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 11 |
Hb_004223_270 |
0.0945009279 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009288_040 |
0.0952959733 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 13 |
Hb_027445_100 |
0.0962488734 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 14 |
Hb_002005_040 |
0.097276904 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_006829_080 |
0.103526256 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 16 |
Hb_000402_020 |
0.1049759171 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001959_060 |
0.1065386334 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003664_030 |
0.1082597957 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 19 |
Hb_000537_070 |
0.1082836142 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 20 |
Hb_025194_090 |
0.109078692 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |