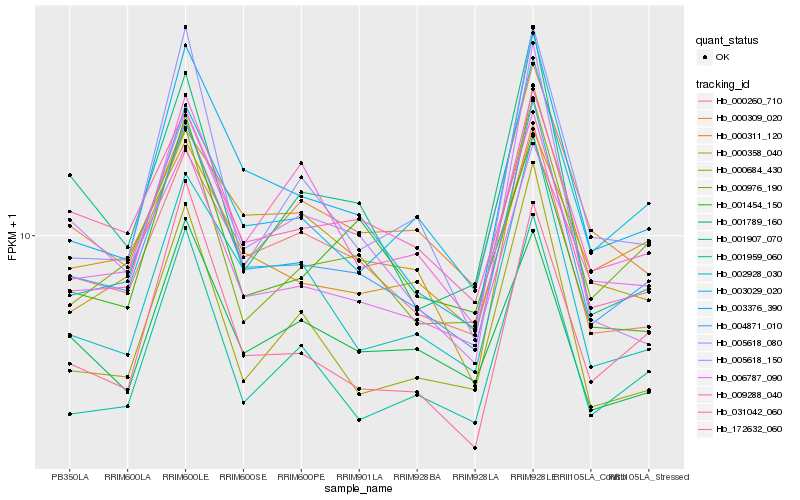
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004871_010 |
0.0 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 2 |
Hb_003376_390 |
0.0682683414 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000260_710 |
0.078972994 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000976_190 |
0.0857771453 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 5 |
Hb_006787_090 |
0.0871831501 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 6 |
Hb_002928_030 |
0.0912200013 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 7 |
Hb_001959_060 |
0.0965855821 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_172632_060 |
0.0984014232 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000358_040 |
0.1007074713 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001789_160 |
0.1044323347 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 11 |
Hb_000311_120 |
0.104677169 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 12 |
Hb_000684_430 |
0.1068883645 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003029_020 |
0.1072507137 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_001907_070 |
0.1082298068 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 15 |
Hb_031042_060 |
0.1085139495 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000309_020 |
0.108547243 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 17 |
Hb_009288_040 |
0.1097152429 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 18 |
Hb_005618_080 |
0.1098629474 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001454_150 |
0.1100746327 |
- |
- |
isoleucyl tRNA synthetase, putative [Ricinus communis] |
| 20 |
Hb_005618_150 |
0.1109349509 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |