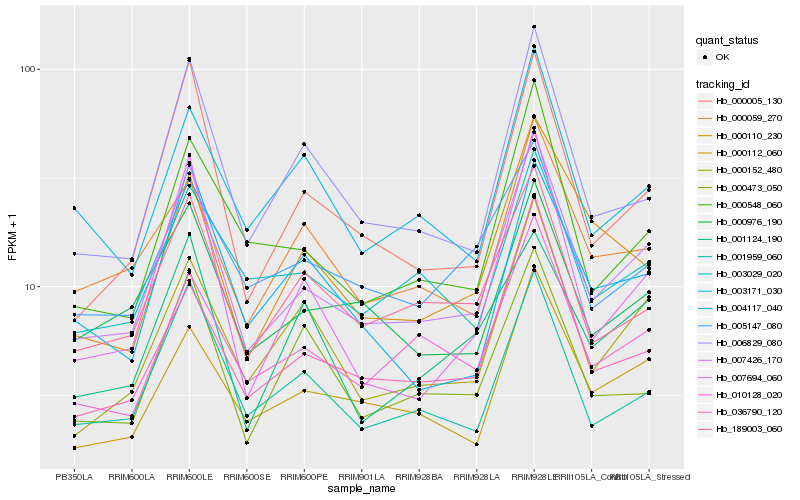
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_170 |
0.0 |
transcription factor |
TF Family: TCP |
- |
| 2 |
Hb_000005_130 |
0.0924401687 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 3 |
Hb_001959_060 |
0.0975445679 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006829_080 |
0.1007657333 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 5 |
Hb_000548_060 |
0.1038943626 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 6 |
Hb_000112_060 |
0.1041843564 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000976_190 |
0.1083158592 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 8 |
Hb_010128_020 |
0.1083464555 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 9 |
Hb_189003_060 |
0.1090680564 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000152_480 |
0.1105389829 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_036790_120 |
0.1145555682 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007694_060 |
0.1175139758 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003029_020 |
0.1219865606 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_000059_270 |
0.1244357492 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 15 |
Hb_000110_230 |
0.1246646818 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_003171_030 |
0.1264377142 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 17 |
Hb_004117_040 |
0.1264429258 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 18 |
Hb_000473_050 |
0.127190851 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 19 |
Hb_005147_080 |
0.1301092727 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001124_190 |
0.1309601682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |