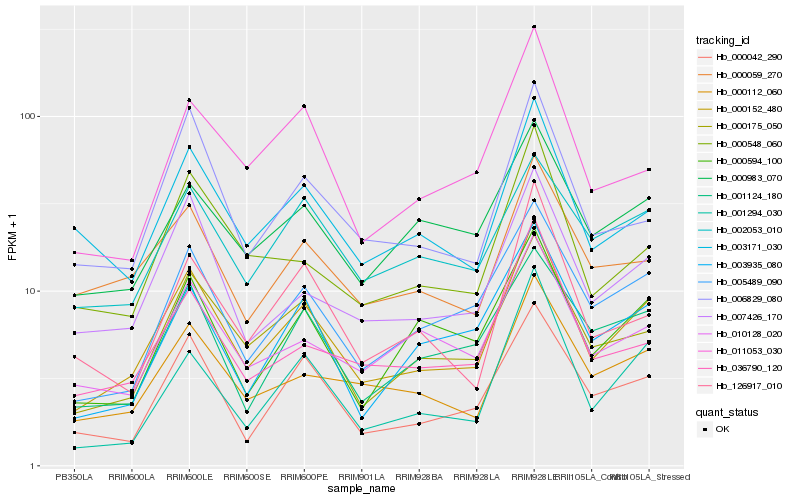
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_480 |
0.0 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000175_050 |
0.0972299787 |
- |
- |
- |
| 3 |
Hb_005489_090 |
0.1035242192 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000112_060 |
0.1059551728 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000042_290 |
0.1064272278 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007426_170 |
0.1105389829 |
transcription factor |
TF Family: TCP |
- |
| 7 |
Hb_036790_120 |
0.1198675494 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003935_080 |
0.1200379187 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000548_060 |
0.1209281711 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 10 |
Hb_000594_100 |
0.1215065171 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006829_080 |
0.1229530189 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 12 |
Hb_003171_030 |
0.1240520211 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 13 |
Hb_011053_030 |
0.1241668269 |
- |
- |
- |
| 14 |
Hb_010128_020 |
0.1245873296 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 15 |
Hb_001294_030 |
0.1260562191 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 16 |
Hb_001124_180 |
0.1263720452 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 17 |
Hb_000983_070 |
0.1281158468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000059_270 |
0.1288983986 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 19 |
Hb_126917_010 |
0.1313429853 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 20 |
Hb_002053_010 |
0.1338951888 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |