| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
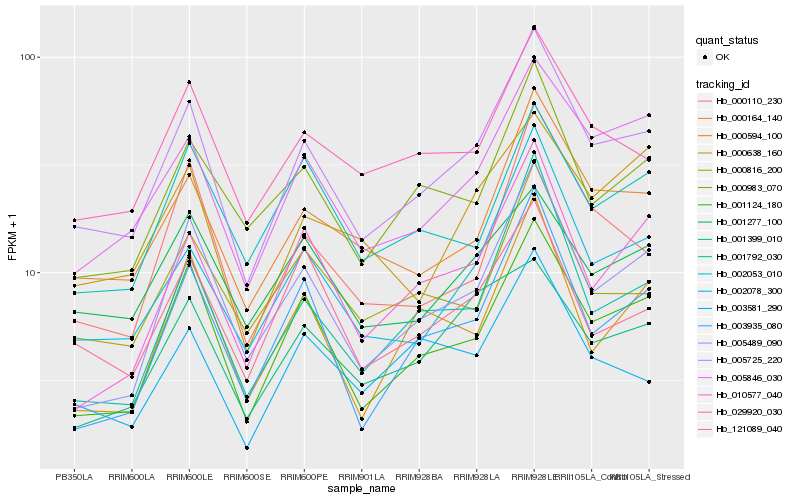
Hb_005725_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 2 |
Hb_000164_140 |
0.0826089002 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_001124_180 |
0.0836253662 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 4 |
Hb_005846_030 |
0.0879343004 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002078_300 |
0.0922637159 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005489_090 |
0.0956510263 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_029920_030 |
0.1013008474 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000983_070 |
0.1050665507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001792_030 |
0.1128536859 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 10 |
Hb_003935_080 |
0.1140161041 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000816_200 |
0.1148631578 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 12 |
Hb_000110_230 |
0.1163662505 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_121089_040 |
0.1163988062 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 14 |
Hb_010577_040 |
0.116738793 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 15 |
Hb_001277_100 |
0.1208631716 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 16 |
Hb_001399_010 |
0.1214585784 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_003581_290 |
0.1235313265 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_002053_010 |
0.1243324636 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000638_160 |
0.1264392641 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 20 |
Hb_000594_100 |
0.1267099297 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |