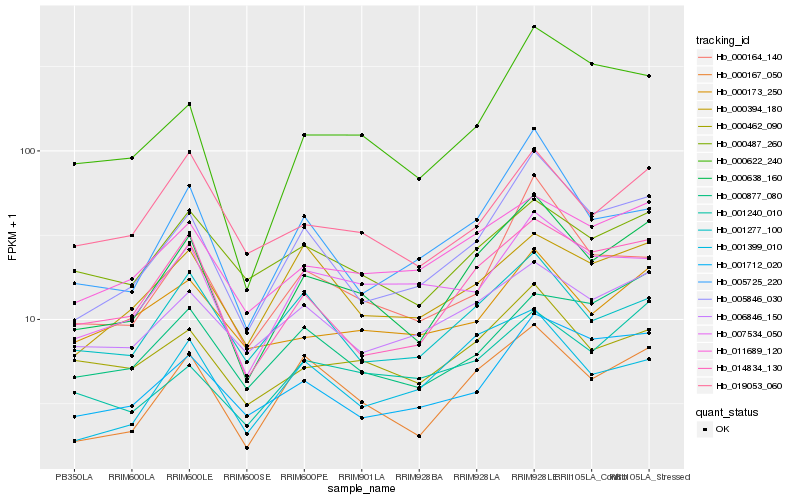
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000638_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 2 |
Hb_000167_050 |
0.0878192649 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 3 |
Hb_005846_030 |
0.0956103475 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 4 |
Hb_014834_130 |
0.0999484895 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_011689_120 |
0.1153631933 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 6 |
Hb_000622_240 |
0.1255718276 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000462_090 |
0.1263006853 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005725_220 |
0.1264392641 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 9 |
Hb_000164_140 |
0.1283746425 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_001712_020 |
0.1303182092 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 11 |
Hb_019053_060 |
0.1330712214 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 12 |
Hb_006846_150 |
0.133217828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001277_100 |
0.1339012873 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 14 |
Hb_001240_010 |
0.1352624112 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 15 |
Hb_007534_050 |
0.1353431363 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001399_010 |
0.1377667498 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000487_260 |
0.138067781 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000394_180 |
0.1423413163 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 19 |
Hb_000173_250 |
0.1431849179 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 20 |
Hb_000877_080 |
0.1433961197 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |