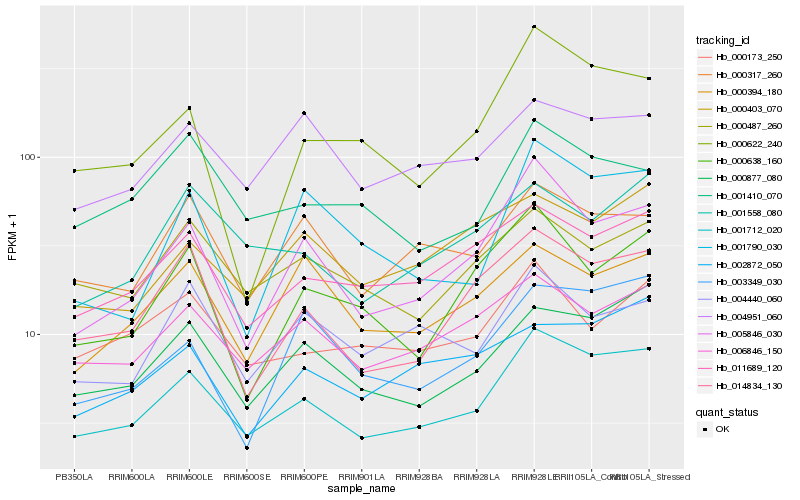
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001712_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 2 |
Hb_000877_080 |
0.0941391855 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 3 |
Hb_005846_030 |
0.0944187945 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011689_120 |
0.1062317268 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 5 |
Hb_006846_150 |
0.1069517838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_014834_130 |
0.1108150205 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000317_260 |
0.1232391513 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_004951_060 |
0.1249983364 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 9 |
Hb_000487_260 |
0.1251667883 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000622_240 |
0.1269840619 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001558_080 |
0.1273837691 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000403_070 |
0.1279684915 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002872_050 |
0.1280551425 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000173_250 |
0.1287236032 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 15 |
Hb_003349_030 |
0.1296896265 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 16 |
Hb_001790_030 |
0.130278359 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000638_160 |
0.1303182092 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 18 |
Hb_000394_180 |
0.1303506603 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 19 |
Hb_001410_070 |
0.1323502464 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 20 |
Hb_004440_060 |
0.1328011753 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |