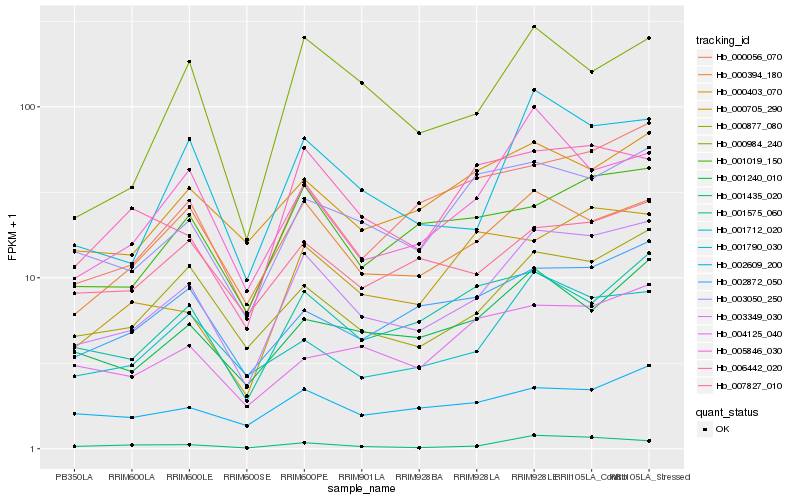
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003349_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 2 |
Hb_000877_080 |
0.1093329586 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 3 |
Hb_001790_030 |
0.120150764 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000056_070 |
0.1215989663 |
- |
- |
- |
| 5 |
Hb_003050_250 |
0.1268500265 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001712_020 |
0.1296896265 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 7 |
Hb_001019_150 |
0.1298231534 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 8 |
Hb_001240_010 |
0.1303607199 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 9 |
Hb_002872_050 |
0.1305024814 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000394_180 |
0.1335326437 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 11 |
Hb_000403_070 |
0.1344853543 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_007827_010 |
0.1346699815 |
- |
- |
PREDICTED: uncharacterized protein C12B10.15c [Jatropha curcas] |
| 13 |
Hb_001435_020 |
0.1363891724 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 14 |
Hb_002609_200 |
0.1388557337 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_001575_060 |
0.1391534706 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000984_240 |
0.1417721948 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 17 |
Hb_005846_030 |
0.1435588438 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006442_020 |
0.1452309762 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 19 |
Hb_004125_040 |
0.1474795437 |
- |
- |
- |
| 20 |
Hb_000705_290 |
0.1495856131 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |