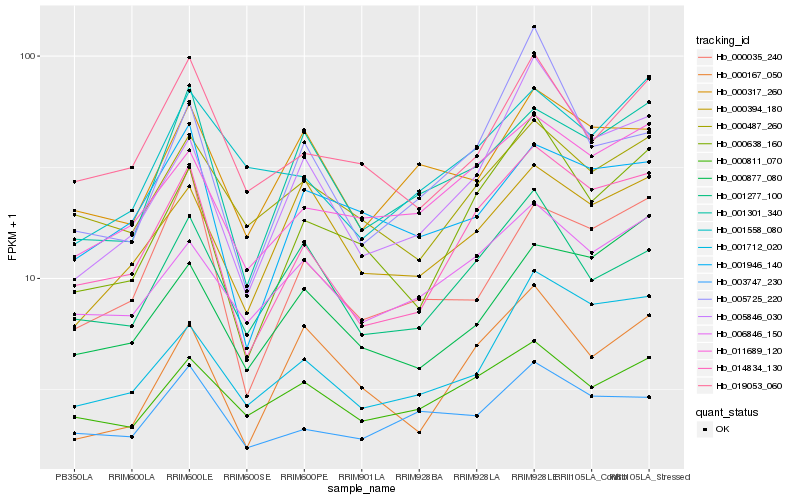
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014834_130 |
0.0 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_000638_160 |
0.0999484895 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 3 |
Hb_001712_020 |
0.1108150205 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 4 |
Hb_005846_030 |
0.113454121 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 5 |
Hb_011689_120 |
0.1153970223 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 6 |
Hb_001301_340 |
0.1219455238 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 7 |
Hb_006846_150 |
0.1261344478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000035_240 |
0.1277164473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000877_080 |
0.1303536291 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 10 |
Hb_001277_100 |
0.1305526115 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 11 |
Hb_000487_260 |
0.1305706701 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 12 |
Hb_019053_060 |
0.1328860539 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 13 |
Hb_001946_140 |
0.1380533497 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000167_050 |
0.1390980925 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 15 |
Hb_005725_220 |
0.1418438149 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 16 |
Hb_001558_080 |
0.1419762206 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000394_180 |
0.1435034717 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 18 |
Hb_000317_260 |
0.1435574437 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_003747_230 |
0.1439529454 |
- |
- |
PREDICTED: uncharacterized protein LOC105630239 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000811_070 |
0.1452632151 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |