| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
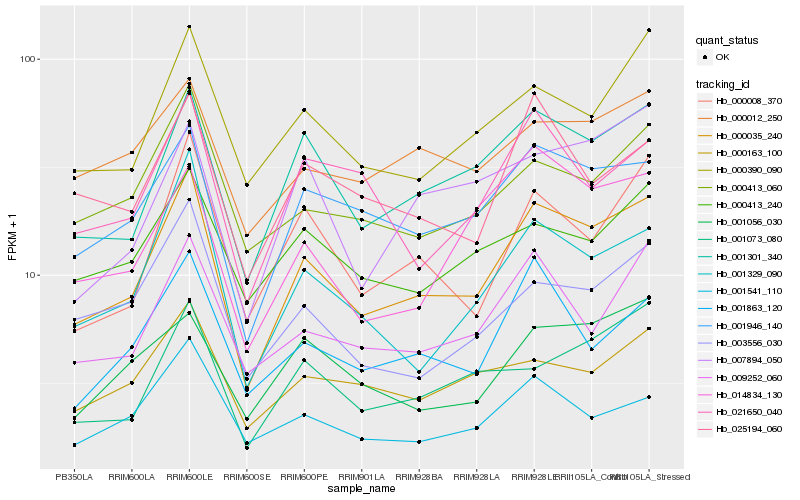
Hb_000035_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001301_340 |
0.0877152267 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 3 |
Hb_001946_140 |
0.0932287901 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_001329_090 |
0.1109407106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000390_090 |
0.1120003063 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 6 |
Hb_000008_370 |
0.1129925684 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 7 |
Hb_009252_060 |
0.1143501026 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 8 |
Hb_001073_080 |
0.1152353858 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000413_060 |
0.1170923868 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 10 |
Hb_001863_120 |
0.1215911191 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003556_030 |
0.1232623604 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 12 |
Hb_000163_100 |
0.1245273193 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 13 |
Hb_001541_110 |
0.126225316 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_001056_030 |
0.1262725125 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 15 |
Hb_025194_060 |
0.1274871381 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_014834_130 |
0.1277164473 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000012_250 |
0.1277213946 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 18 |
Hb_000413_240 |
0.1279326375 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_021650_040 |
0.1280259068 |
- |
- |
EG2771 [Manihot esculenta] |
| 20 |
Hb_007894_050 |
0.129994091 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |