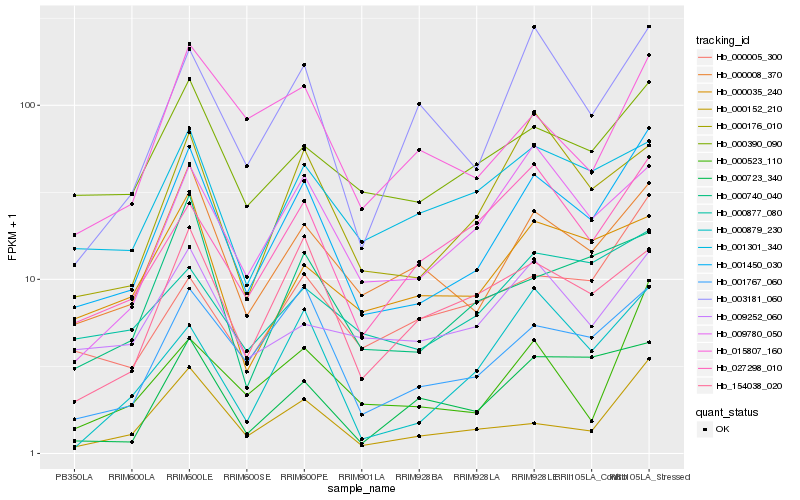
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001450_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 2 |
Hb_000008_370 |
0.1298594283 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 3 |
Hb_001767_060 |
0.1331786408 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000390_090 |
0.1493343303 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 5 |
Hb_000152_210 |
0.1507589412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000176_010 |
0.1589173678 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003181_060 |
0.1589593868 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 8 |
Hb_009780_050 |
0.1601897485 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000740_040 |
0.1614879613 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 10 |
Hb_000523_110 |
0.1648872018 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 11 |
Hb_154038_020 |
0.1651701132 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 12 |
Hb_027298_010 |
0.1655518244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001301_340 |
0.1713148438 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 14 |
Hb_000877_080 |
0.1723414639 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 15 |
Hb_000879_230 |
0.1728182857 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 16 |
Hb_015807_160 |
0.1730820533 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 17 |
Hb_000005_300 |
0.1734949666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000035_240 |
0.1736294998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009252_060 |
0.1740980916 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 20 |
Hb_000723_340 |
0.1750110771 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |