| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
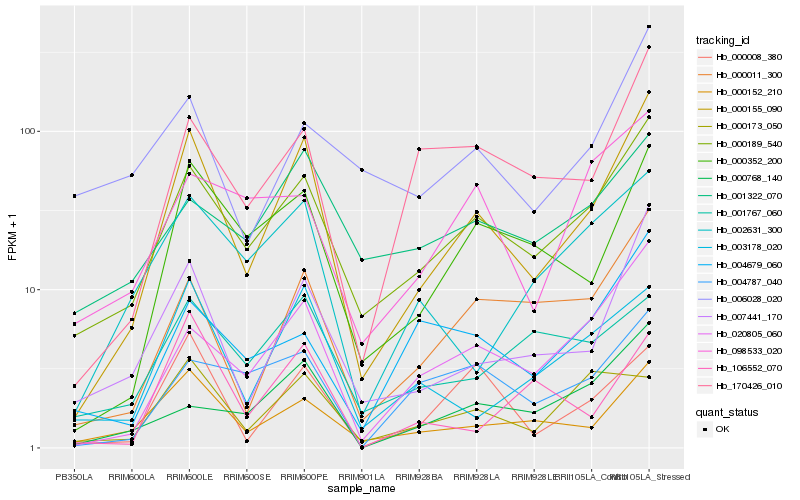
Hb_000155_090 |
0.0 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 2 |
Hb_000189_540 |
0.1478414384 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007441_170 |
0.1527096772 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 4 |
Hb_020805_060 |
0.1699944379 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 5 |
Hb_000152_210 |
0.1749715976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000352_200 |
0.1767253864 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000011_300 |
0.1822194198 |
- |
- |
PREDICTED: proteasome subunit alpha type-5 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_170426_010 |
0.1985761234 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 9 |
Hb_000008_380 |
0.2037095349 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_003178_020 |
0.2039651244 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000768_140 |
0.2217007654 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002631_300 |
0.2265976032 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 13 |
Hb_004787_040 |
0.228220323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004679_060 |
0.2387304844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006028_020 |
0.2422713644 |
- |
- |
PREDICTED: 60S ribosomal protein L38-like [Cicer arietinum] |
| 16 |
Hb_000173_050 |
0.2433288951 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 17 |
Hb_001322_070 |
0.2452328513 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_106552_070 |
0.245853314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_098533_020 |
0.247817451 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 20 |
Hb_001767_060 |
0.2491629579 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |