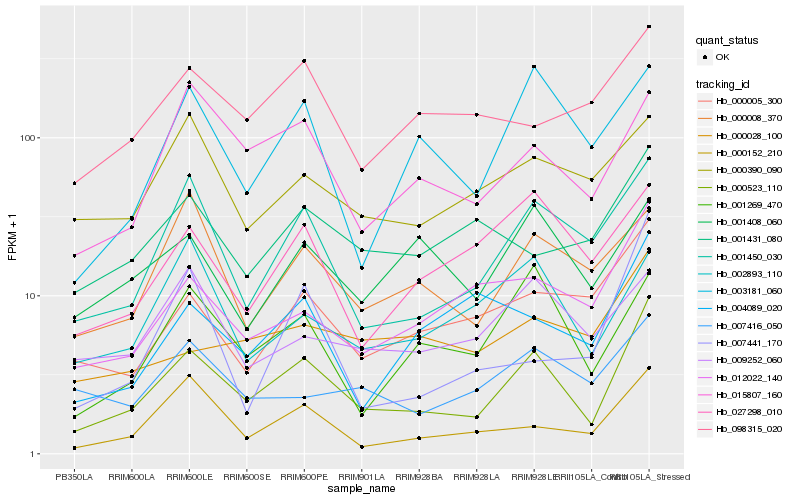
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000523_110 |
0.0 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 2 |
Hb_001450_030 |
0.1648872018 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 3 |
Hb_000152_210 |
0.1896811074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012022_140 |
0.1921766955 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 5 |
Hb_001269_470 |
0.1940947231 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 6 |
Hb_000005_300 |
0.196412261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_015807_160 |
0.1966082003 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 8 |
Hb_003181_060 |
0.2062303497 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 9 |
Hb_027298_010 |
0.2115810905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007416_050 |
0.2133576322 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 11 |
Hb_000008_370 |
0.214043455 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 12 |
Hb_000028_100 |
0.2179464935 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 13 |
Hb_001431_080 |
0.2187292632 |
- |
- |
40S ribosomal protein S11, putative [Ricinus communis] |
| 14 |
Hb_001408_060 |
0.2188842231 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 15 |
Hb_004089_020 |
0.2189584882 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 16 |
Hb_098315_020 |
0.2202162616 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 17 |
Hb_007441_170 |
0.2202982907 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 18 |
Hb_002893_110 |
0.2218601855 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000390_090 |
0.2226133536 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 20 |
Hb_009252_060 |
0.2227826843 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |