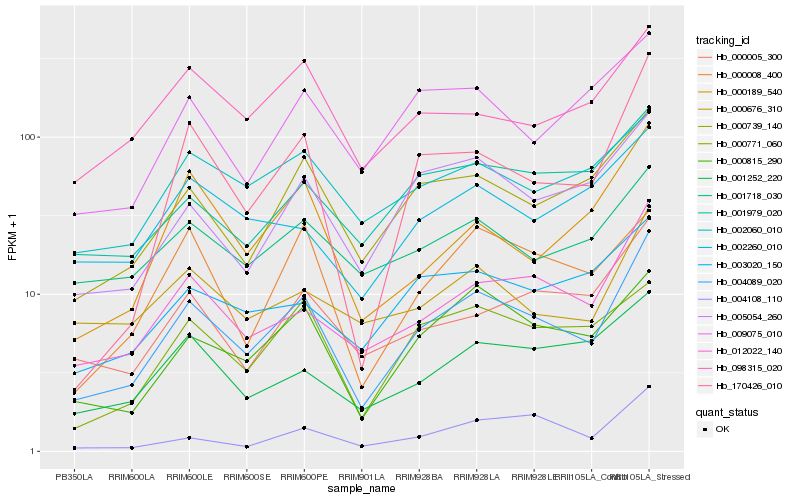
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004089_020 |
0.0 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 2 |
Hb_012022_140 |
0.121417402 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 3 |
Hb_000815_290 |
0.1249596888 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000739_140 |
0.1256444263 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 5 |
Hb_005054_260 |
0.134866069 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 6 |
Hb_170426_010 |
0.1374627411 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 7 |
Hb_000005_300 |
0.1446789593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000771_060 |
0.147353832 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 9 |
Hb_001979_020 |
0.15020122 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 10 |
Hb_001252_220 |
0.1513880136 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 11 |
Hb_000676_310 |
0.1537874537 |
- |
- |
BnaC09g40680D [Brassica napus] |
| 12 |
Hb_000189_540 |
0.1556752004 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000008_400 |
0.1561540323 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 14 |
Hb_003020_150 |
0.1564604922 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 15 |
Hb_009075_010 |
0.1566510089 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 16 |
Hb_001718_030 |
0.1598181746 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002060_010 |
0.1607347359 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004108_110 |
0.1611674114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_098315_020 |
0.162609047 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 20 |
Hb_002260_010 |
0.1636057303 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |