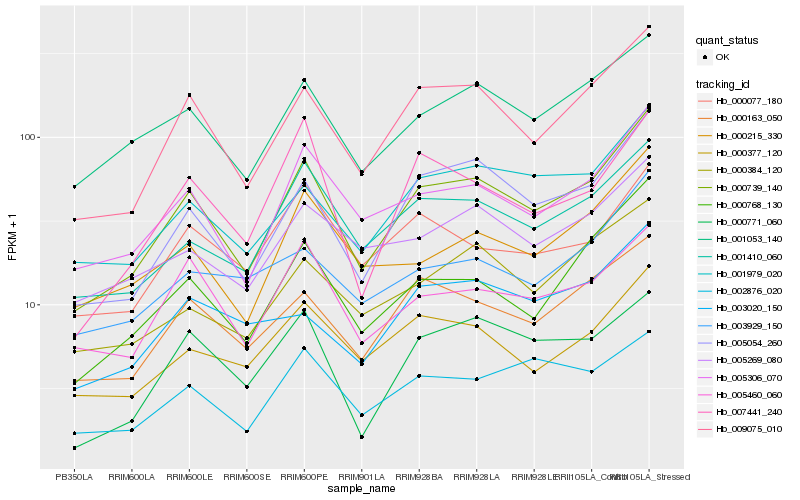
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_140 |
0.0 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 2 |
Hb_009075_010 |
0.0638372924 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 3 |
Hb_005054_260 |
0.0752162047 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 4 |
Hb_005306_070 |
0.081512271 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 5 |
Hb_000768_130 |
0.0927854822 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 6 |
Hb_001053_140 |
0.09480202 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 7 |
Hb_001410_060 |
0.0965395131 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 8 |
Hb_000163_050 |
0.0965545999 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 9 |
Hb_001979_020 |
0.0997500319 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 10 |
Hb_000215_330 |
0.1065770316 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 11 |
Hb_000377_120 |
0.1101622999 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000384_120 |
0.1135074346 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_005269_080 |
0.1146718702 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_005460_060 |
0.1166597646 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 15 |
Hb_003929_150 |
0.1184879061 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 16 |
Hb_007441_240 |
0.1207057238 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 17 |
Hb_000771_060 |
0.1215838539 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 18 |
Hb_002876_020 |
0.1217530955 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003020_150 |
0.1226984608 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 20 |
Hb_000077_180 |
0.1230454363 |
- |
- |
- |