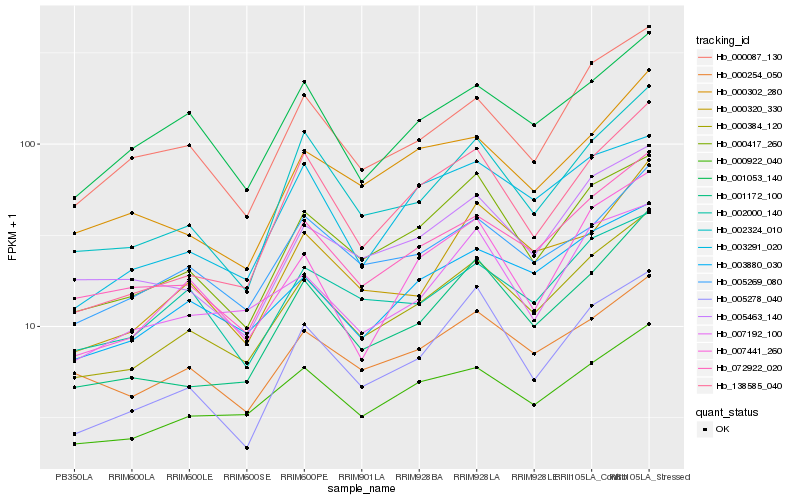
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000384_120 |
0.0 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_072922_020 |
0.0591047997 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 3 |
Hb_000417_260 |
0.0686941531 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_003880_030 |
0.0696665208 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000922_040 |
0.0714703597 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 6 |
Hb_005269_080 |
0.072860474 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_002324_010 |
0.0763923767 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 8 |
Hb_001172_100 |
0.0781173299 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000087_130 |
0.0795322869 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 10 |
Hb_005463_140 |
0.0817824572 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 11 |
Hb_000254_050 |
0.0843027056 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 12 |
Hb_000320_330 |
0.0851027833 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 13 |
Hb_002000_140 |
0.0872576316 |
- |
- |
PREDICTED: thioredoxin Y2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001053_140 |
0.0883630905 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 15 |
Hb_007192_100 |
0.0890961363 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 16 |
Hb_138585_040 |
0.089196124 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 17 |
Hb_007441_260 |
0.0894363951 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_005278_040 |
0.0894714406 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000302_280 |
0.0895412501 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 20 |
Hb_003291_020 |
0.0897264905 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |