| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
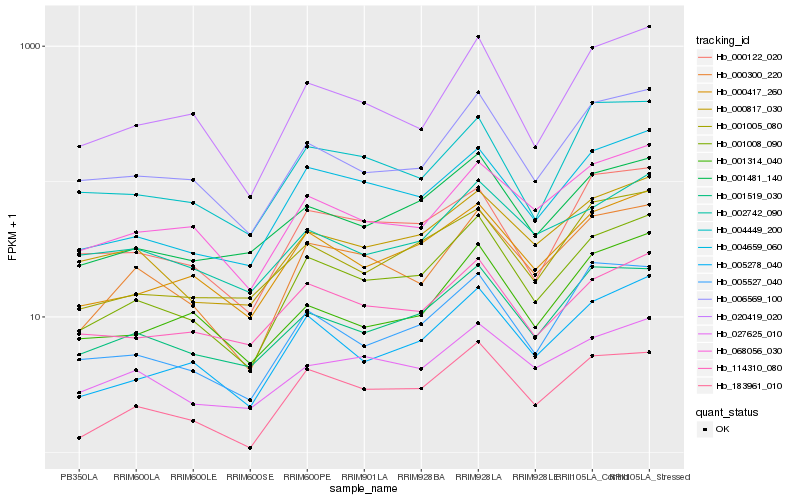
Hb_001008_090 |
0.0 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 2 |
Hb_005278_040 |
0.073763881 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000417_260 |
0.0749510162 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_004659_060 |
0.0770520222 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 5 |
Hb_006569_100 |
0.0774611074 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |
| 6 |
Hb_000300_220 |
0.0774660443 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 7 |
Hb_183961_010 |
0.0858656198 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 8 |
Hb_001481_140 |
0.0865618373 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 9 |
Hb_001519_030 |
0.0874471653 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 10 |
Hb_020419_020 |
0.0899462473 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_000817_030 |
0.0937090015 |
- |
- |
mak, putative [Ricinus communis] |
| 12 |
Hb_068056_030 |
0.0956118564 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 13 |
Hb_005527_040 |
0.0959934211 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 14 |
Hb_114310_080 |
0.0998922772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001005_080 |
0.0999757579 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000122_020 |
0.1005304579 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |
| 17 |
Hb_027625_010 |
0.1030901227 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 18 |
Hb_001314_040 |
0.1076281062 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 19 |
Hb_002742_090 |
0.1087768513 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 20 |
Hb_004449_200 |
0.109692648 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Populus euphratica] |