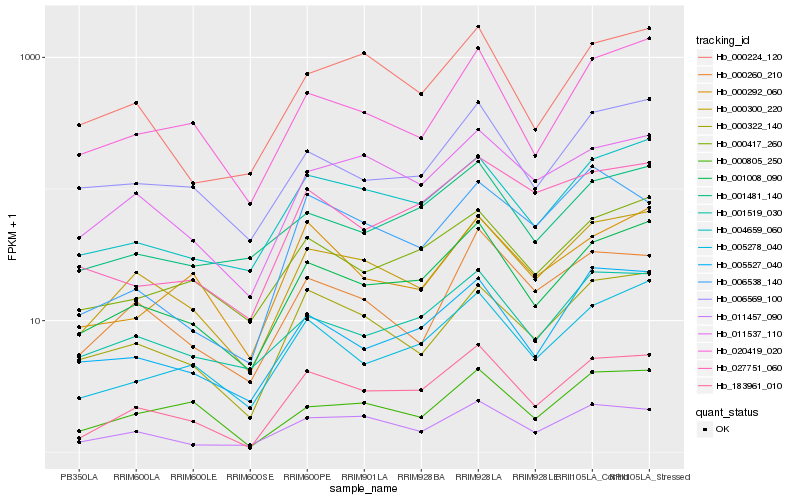
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183961_010 |
0.0 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 2 |
Hb_001008_090 |
0.0858656198 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 3 |
Hb_000300_220 |
0.0938407243 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 4 |
Hb_005278_040 |
0.1180098625 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 5 |
Hb_011457_090 |
0.1180103152 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 6 |
Hb_011537_110 |
0.1217137345 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 7 |
Hb_004659_060 |
0.121898846 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 8 |
Hb_000322_140 |
0.1258585315 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 9 |
Hb_000417_260 |
0.1320136151 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_005527_040 |
0.1325957695 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 11 |
Hb_000260_210 |
0.1365960197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_027751_060 |
0.139099311 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 13 |
Hb_001519_030 |
0.1393289062 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000224_120 |
0.1422285599 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 15 |
Hb_020419_020 |
0.1433069611 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 16 |
Hb_006538_140 |
0.1438023366 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 17 |
Hb_006569_100 |
0.1452652515 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |
| 18 |
Hb_001481_140 |
0.1455755169 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 19 |
Hb_000292_060 |
0.1467230606 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_000805_250 |
0.1486052812 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |