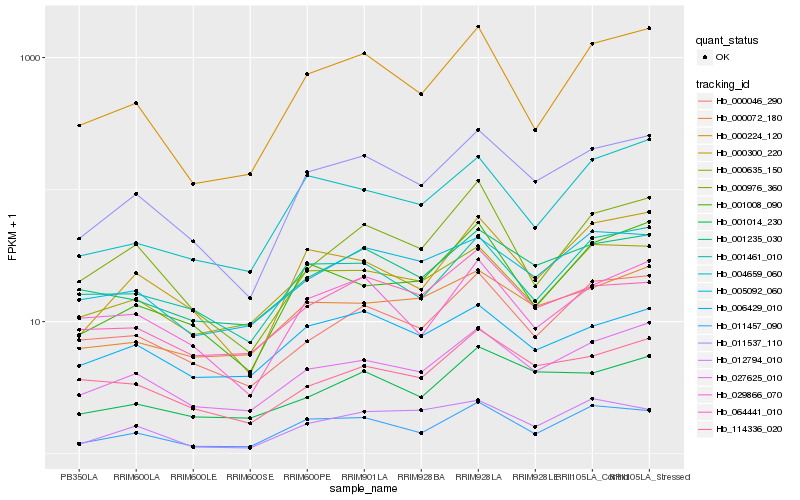
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_110 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 2 |
Hb_027625_010 |
0.0794296635 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 3 |
Hb_011457_090 |
0.0819241268 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000300_220 |
0.0883148258 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 5 |
Hb_001235_030 |
0.1024681837 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 6 |
Hb_000224_120 |
0.1033613021 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 7 |
Hb_000046_290 |
0.1044286801 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 8 |
Hb_029866_070 |
0.1045642117 |
- |
- |
PREDICTED: myosin IB heavy chain [Jatropha curcas] |
| 9 |
Hb_000072_180 |
0.1064460599 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 10 |
Hb_005092_060 |
0.107120352 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 11 |
Hb_001014_230 |
0.1095746325 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 12 |
Hb_001008_090 |
0.1105469411 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 13 |
Hb_001461_010 |
0.1123023583 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Pyrus x bretschneideri] |
| 14 |
Hb_000635_150 |
0.113045426 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 15 |
Hb_114336_020 |
0.1136219513 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 16 |
Hb_004659_060 |
0.1155298888 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 17 |
Hb_012794_010 |
0.1161803742 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 18 |
Hb_006429_010 |
0.116333656 |
- |
- |
PREDICTED: uncharacterized protein LOC105639724 [Jatropha curcas] |
| 19 |
Hb_064441_010 |
0.1164432229 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000976_360 |
0.1175418191 |
- |
- |
PREDICTED: uncharacterized protein LOC105646237 [Jatropha curcas] |