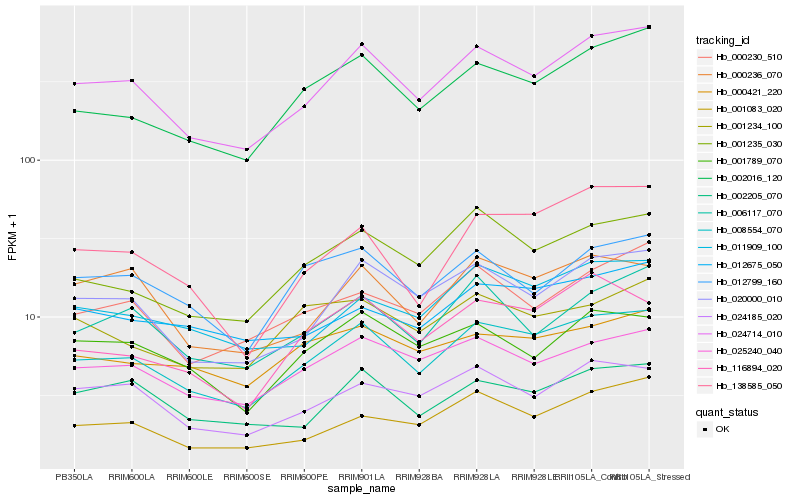
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008554_070 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 3-2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_024714_010 |
0.044403366 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_002016_120 |
0.0690129831 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940 [Jatropha curcas] |
| 4 |
Hb_020000_010 |
0.0794027373 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 5 |
Hb_001235_030 |
0.0799450862 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 6 |
Hb_025240_040 |
0.0820401811 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |
| 7 |
Hb_011909_100 |
0.0870646052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_116894_020 |
0.0882230582 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 9 |
Hb_000421_220 |
0.0941156669 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_024185_020 |
0.0948105745 |
- |
- |
- |
| 11 |
Hb_138585_050 |
0.0948605589 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 12 |
Hb_012799_160 |
0.0953412361 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1-like [Jatropha curcas] |
| 13 |
Hb_006117_070 |
0.0961460674 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000236_070 |
0.0961534468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002205_070 |
0.0968285114 |
- |
- |
PREDICTED: tRNA (guanine(26)-N(2))-dimethyltransferase isoform X1 [Jatropha curcas] |
| 16 |
Hb_001083_020 |
0.0971822937 |
- |
- |
- |
| 17 |
Hb_012675_050 |
0.0990228943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000230_510 |
0.0992038631 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 19 |
Hb_001789_070 |
0.0992334265 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 12 [Jatropha curcas] |
| 20 |
Hb_001234_100 |
0.1002952999 |
- |
- |
PREDICTED: eyes absent homolog 2 isoform X1 [Jatropha curcas] |