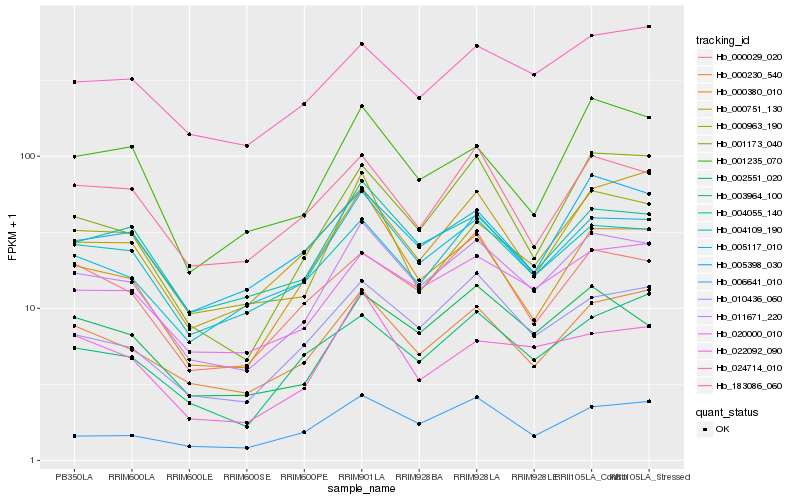
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_220 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005117_010 |
0.0760760866 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_020000_010 |
0.0814764209 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 4 |
Hb_000230_540 |
0.0858728569 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 5 |
Hb_004055_140 |
0.0865526027 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_010436_060 |
0.0883658334 |
- |
- |
hypothetical protein CISIN_1g0003801mg, partial [Citrus sinensis] |
| 7 |
Hb_002551_020 |
0.0927959389 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004109_190 |
0.0937719705 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 25-like isoform X2 [Populus euphratica] |
| 9 |
Hb_005398_030 |
0.1016219232 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase 1-like [Jatropha curcas] |
| 10 |
Hb_000963_190 |
0.1020067205 |
- |
- |
Glutamine amidotransferase subunit pdxT, putative [Ricinus communis] |
| 11 |
Hb_024714_010 |
0.1060105329 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_006641_010 |
0.10624709 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 13 |
Hb_000029_020 |
0.1082068462 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 14 |
Hb_000380_010 |
0.1087090569 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X3 [Jatropha curcas] |
| 15 |
Hb_001235_070 |
0.1090165193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_022092_090 |
0.1101041508 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 17 |
Hb_183086_060 |
0.1101075785 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |
| 18 |
Hb_001173_040 |
0.1112731599 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_003964_100 |
0.1115428971 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 20 |
Hb_000751_130 |
0.1116144417 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |