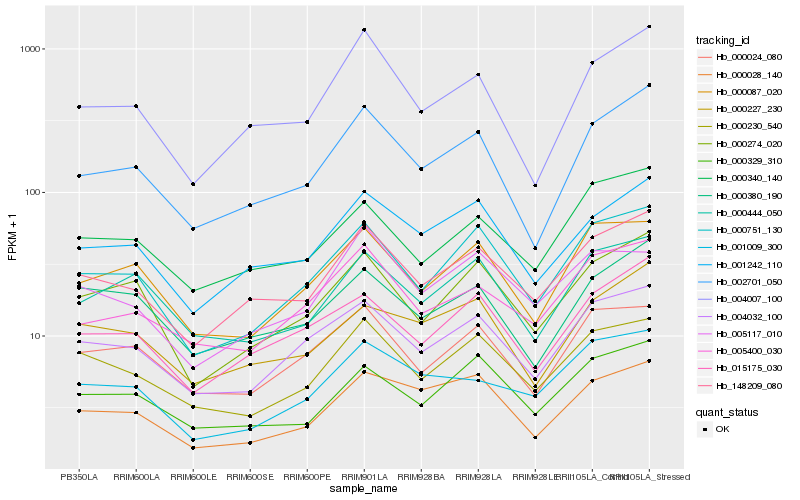
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148209_080 |
0.0 |
- |
- |
MED32, putative [Theobroma cacao] |
| 2 |
Hb_001242_110 |
0.068096206 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 3 |
Hb_000340_140 |
0.0791070725 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Vitis vinifera] |
| 4 |
Hb_000230_540 |
0.0800604169 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 5 |
Hb_004032_100 |
0.0809765019 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like [Jatropha curcas] |
| 6 |
Hb_000751_130 |
0.0828674396 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 7 |
Hb_015175_030 |
0.0832006609 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 8 |
Hb_000274_020 |
0.0884943223 |
- |
- |
PREDICTED: uncharacterized protein LOC105630805 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000329_310 |
0.0887891008 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005400_030 |
0.089017051 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 11 |
Hb_002701_050 |
0.0899502057 |
- |
- |
PREDICTED: 60S ribosomal protein L7a-like [Jatropha curcas] |
| 12 |
Hb_004007_100 |
0.0908586991 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 13 |
Hb_001009_300 |
0.0968212567 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 14 |
Hb_005117_010 |
0.0982222945 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000024_080 |
0.0994163618 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000380_190 |
0.0999184434 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_000227_230 |
0.1002991064 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 18 |
Hb_000087_020 |
0.1018650332 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase 2, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000028_140 |
0.1022874701 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 20 |
Hb_000444_050 |
0.1033437663 |
transcription factor |
TF Family: HMG |
PREDICTED: uncharacterized protein LOC105648212 isoform X2 [Jatropha curcas] |