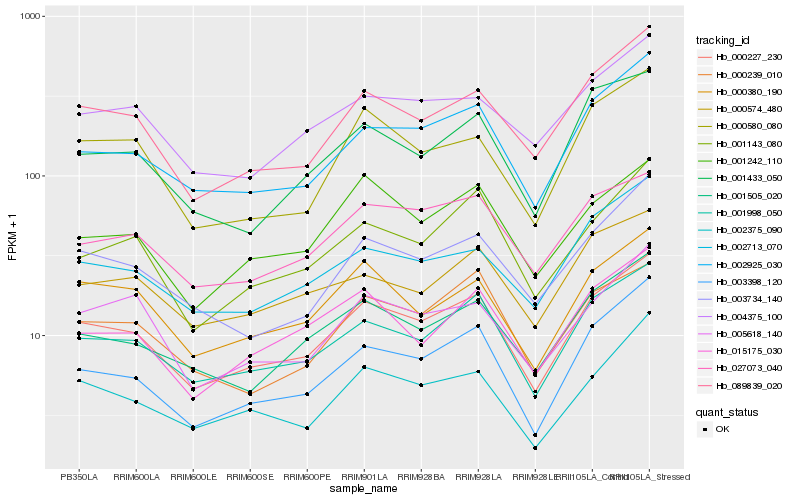
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_230 |
0.0 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 2 |
Hb_001998_050 |
0.0483249402 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_002925_030 |
0.0634908676 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 4 |
Hb_089839_020 |
0.0680335227 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 5 |
Hb_003398_120 |
0.068658753 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 6 |
Hb_001505_020 |
0.0695137741 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 9 [Jatropha curcas] |
| 7 |
Hb_000380_190 |
0.07261918 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 8 |
Hb_002375_090 |
0.0744484767 |
- |
- |
- |
| 9 |
Hb_001143_080 |
0.0767440889 |
- |
- |
PREDICTED: F-box protein 7 [Jatropha curcas] |
| 10 |
Hb_027073_040 |
0.0768562504 |
- |
- |
TGF-beta-inducible nuclear protein, putative [Ricinus communis] |
| 11 |
Hb_003734_140 |
0.0778914022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002713_070 |
0.079449653 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 13 |
Hb_015175_030 |
0.0801521508 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 14 |
Hb_004375_100 |
0.081314776 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 15 |
Hb_001242_110 |
0.0818666596 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 16 |
Hb_005618_140 |
0.0821815067 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 17 |
Hb_001433_050 |
0.0830992519 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 18 |
Hb_000239_010 |
0.0836916088 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000574_480 |
0.0846584026 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 20 |
Hb_000580_080 |
0.0849573042 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |