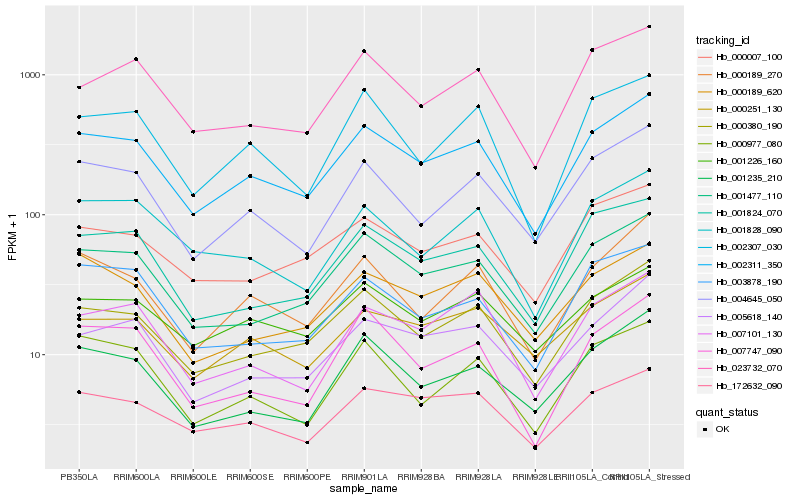
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_350 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 2 |
Hb_000380_190 |
0.0511772009 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 3 |
Hb_001477_110 |
0.0615603045 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 4 |
Hb_004645_050 |
0.068683074 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 5 |
Hb_000189_270 |
0.0727598379 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 6 |
Hb_005618_140 |
0.074958232 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 7 |
Hb_001824_070 |
0.0752053973 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 8 |
Hb_000007_100 |
0.0760896211 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 9 |
Hb_172632_090 |
0.0775614317 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 10 |
Hb_001235_210 |
0.0781186343 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_023732_070 |
0.0793666672 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 12 |
Hb_000251_130 |
0.080791051 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 13 |
Hb_002307_030 |
0.0810884936 |
- |
- |
40S ribosomal protein S15D [Hevea brasiliensis] |
| 14 |
Hb_001828_090 |
0.0824675088 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 15 |
Hb_001226_160 |
0.0832690903 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003878_190 |
0.085261432 |
- |
- |
hypothetical protein POPTR_0011s11040g [Populus trichocarpa] |
| 17 |
Hb_000977_080 |
0.0865946387 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 18 |
Hb_000189_620 |
0.0870306785 |
- |
- |
hypothetical protein JCGZ_23450 [Jatropha curcas] |
| 19 |
Hb_007747_090 |
0.0879524687 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 20 |
Hb_007101_130 |
0.0879918569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |