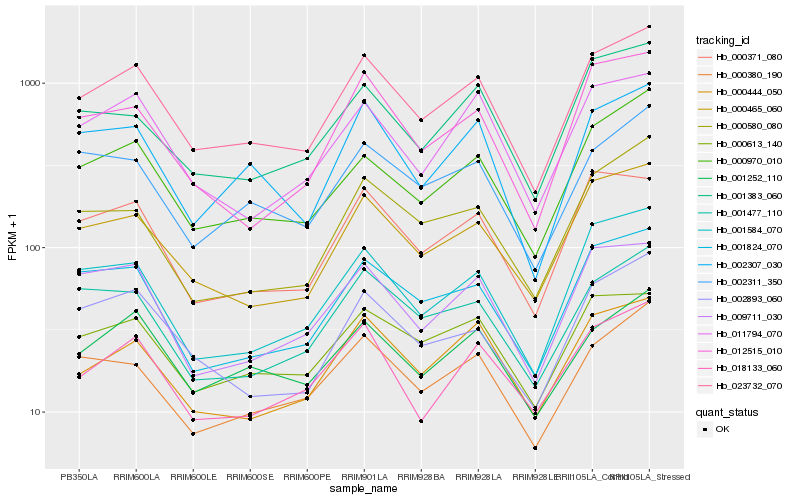
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023732_070 |
0.0 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 2 |
Hb_000465_060 |
0.0588808076 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000970_010 |
0.0663642584 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 4 |
Hb_001824_070 |
0.0680536942 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 5 |
Hb_000444_050 |
0.0696334568 |
transcription factor |
TF Family: HMG |
PREDICTED: uncharacterized protein LOC105648212 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000371_080 |
0.0713567614 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 7 |
Hb_001584_070 |
0.072673298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002893_060 |
0.072677917 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 9 |
Hb_000380_190 |
0.0754679302 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 10 |
Hb_001477_110 |
0.0767839428 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 11 |
Hb_001252_110 |
0.0774737622 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_018133_060 |
0.0785961479 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit RPA12-like [Jatropha curcas] |
| 13 |
Hb_009711_030 |
0.078933068 |
- |
- |
PREDICTED: uncharacterized protein LOC105647792 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002311_350 |
0.0793666672 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 15 |
Hb_000613_140 |
0.079372916 |
- |
- |
PREDICTED: indole-3-glycerol phosphate synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002307_030 |
0.0794936412 |
- |
- |
40S ribosomal protein S15D [Hevea brasiliensis] |
| 17 |
Hb_011794_070 |
0.0810175237 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 18 |
Hb_012515_010 |
0.0830358389 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |
| 19 |
Hb_000580_080 |
0.0831102406 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 20 |
Hb_001383_060 |
0.0831251243 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |