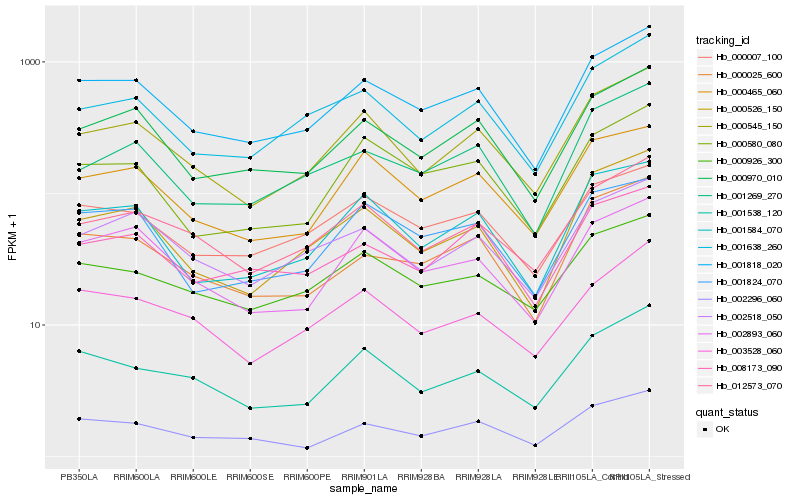
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001818_020 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |
| 2 |
Hb_000970_010 |
0.0486612389 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 3 |
Hb_000545_150 |
0.0652294596 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 4 |
Hb_001584_070 |
0.0685016681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000025_600 |
0.069112533 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000007_100 |
0.0712382918 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 7 |
Hb_001638_260 |
0.0719394281 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1 [Vitis vinifera] |
| 8 |
Hb_002296_060 |
0.0721222159 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_012573_070 |
0.0724445569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002893_060 |
0.0749107148 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 11 |
Hb_000465_060 |
0.0749147279 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002518_050 |
0.0749676653 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 13 |
Hb_000526_150 |
0.0752433678 |
- |
- |
hypothetical protein POPTR_0019s10790g, partial [Populus trichocarpa] |
| 14 |
Hb_003528_060 |
0.0776200516 |
- |
- |
PREDICTED: uncharacterized protein LOC105638256 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001538_120 |
0.0777081103 |
- |
- |
PREDICTED: FAD-linked sulfhydryl oxidase ERV1 [Jatropha curcas] |
| 16 |
Hb_001269_270 |
0.0783102221 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 17 |
Hb_000580_080 |
0.0787773615 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 18 |
Hb_001824_070 |
0.0790529461 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 19 |
Hb_008173_090 |
0.0792807304 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 20 |
Hb_000926_300 |
0.0794349596 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |