| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
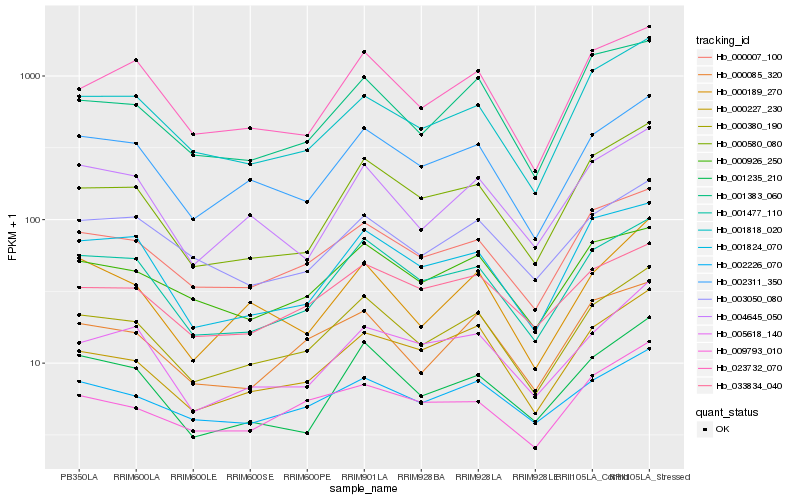
Hb_000380_190 |
0.0 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 2 |
Hb_002311_350 |
0.0511772009 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 3 |
Hb_001477_110 |
0.0539713244 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 4 |
Hb_000007_100 |
0.0559448306 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 5 |
Hb_000227_230 |
0.07261918 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 6 |
Hb_000085_320 |
0.0750038908 |
- |
- |
Uncharacterized protein TCM_030749 [Theobroma cacao] |
| 7 |
Hb_023732_070 |
0.0754679302 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 8 |
Hb_000926_250 |
0.0768432138 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 9 |
Hb_001824_070 |
0.0793330737 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 10 |
Hb_005618_140 |
0.0801214351 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 11 |
Hb_033834_040 |
0.0811992304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000580_080 |
0.0818712117 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 13 |
Hb_001235_210 |
0.082104221 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_001818_020 |
0.0822843672 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |
| 15 |
Hb_003050_080 |
0.0822947867 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 16 |
Hb_002226_070 |
0.0837538806 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001383_060 |
0.0841188036 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 18 |
Hb_000189_270 |
0.0848292554 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 19 |
Hb_004645_050 |
0.086218246 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 20 |
Hb_009793_010 |
0.0868403936 |
- |
- |
hypothetical protein JCGZ_17222 [Jatropha curcas] |