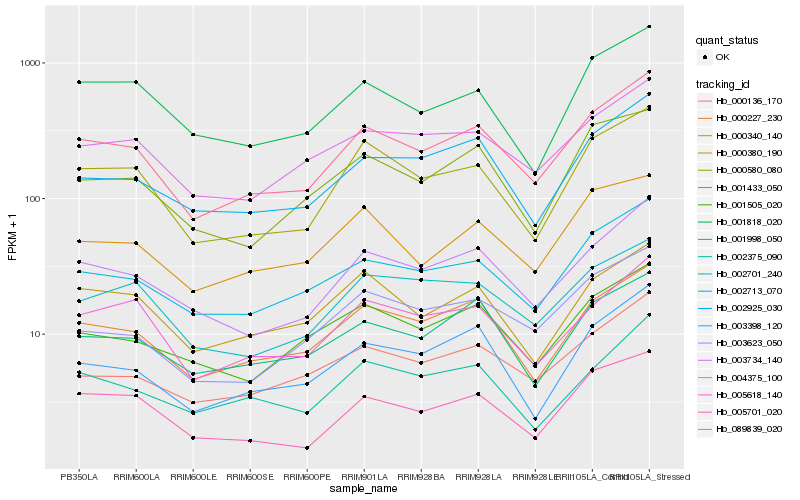
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089839_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 2 |
Hb_003734_140 |
0.0566039117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002713_070 |
0.0647426983 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 4 |
Hb_000227_230 |
0.0680335227 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 5 |
Hb_000580_080 |
0.0689034327 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 6 |
Hb_002925_030 |
0.076175747 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 7 |
Hb_000136_170 |
0.0792855786 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 8 |
Hb_004375_100 |
0.079831601 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 9 |
Hb_005618_140 |
0.0820743326 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 10 |
Hb_003398_120 |
0.0837156834 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 11 |
Hb_001998_050 |
0.0854903237 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_001505_020 |
0.0870501883 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 9 [Jatropha curcas] |
| 13 |
Hb_001433_050 |
0.0876568417 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 14 |
Hb_005701_020 |
0.0888683131 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 15 |
Hb_003623_050 |
0.0889555084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000340_140 |
0.0899112301 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Vitis vinifera] |
| 17 |
Hb_001818_020 |
0.0900455107 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |
| 18 |
Hb_002375_090 |
0.0917564121 |
- |
- |
- |
| 19 |
Hb_002701_240 |
0.0923423626 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 20 |
Hb_000380_190 |
0.0929792454 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |