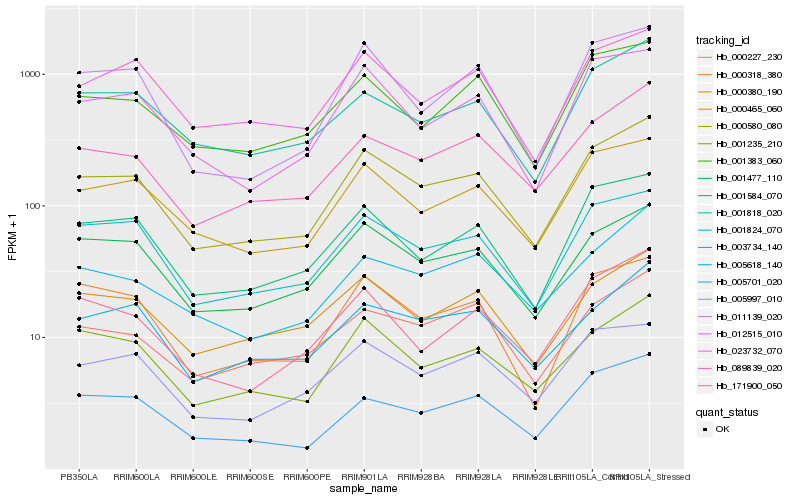
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000580_080 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 2 |
Hb_000465_060 |
0.0665433386 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_089839_020 |
0.0689034327 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 4 |
Hb_001824_070 |
0.0699910759 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 5 |
Hb_001584_070 |
0.0711574637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001235_210 |
0.0737088461 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_001477_110 |
0.0743794522 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 8 |
Hb_012515_010 |
0.0785698029 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |
| 9 |
Hb_001818_020 |
0.0787773615 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |
| 10 |
Hb_005701_020 |
0.0796875771 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 11 |
Hb_000380_190 |
0.0818712117 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 12 |
Hb_023732_070 |
0.0831102406 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 13 |
Hb_005618_140 |
0.0836376474 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 14 |
Hb_005997_010 |
0.0836718067 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 15 |
Hb_171900_050 |
0.0848570873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000227_230 |
0.0849573042 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 17 |
Hb_001383_060 |
0.085208094 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 18 |
Hb_003734_140 |
0.085430738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011139_020 |
0.0864541649 |
- |
- |
Mitochondrial import receptor subunit TOM7-1, putative [Ricinus communis] |
| 20 |
Hb_000318_380 |
0.0870510262 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 3-like [Jatropha curcas] |