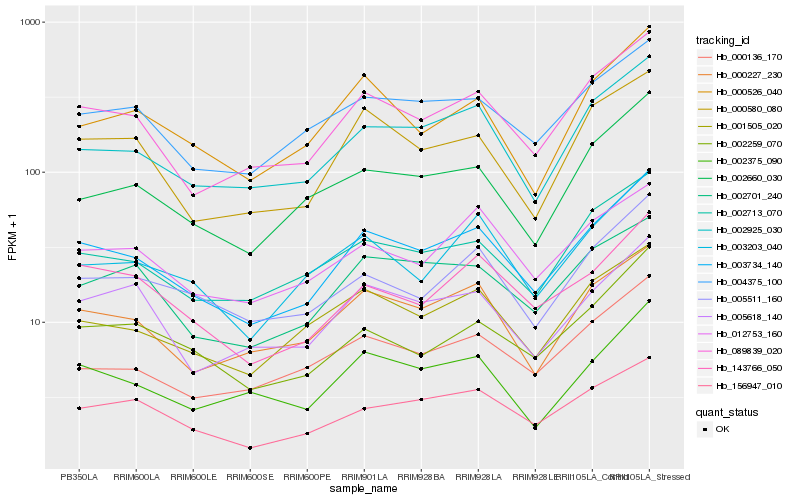
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003734_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_089839_020 |
0.0566039117 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 3 |
Hb_002925_030 |
0.0718033971 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 4 |
Hb_002713_070 |
0.0768235838 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 5 |
Hb_000227_230 |
0.0778914022 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 6 |
Hb_001505_020 |
0.0802184225 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 9 [Jatropha curcas] |
| 7 |
Hb_004375_100 |
0.0812065844 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 8 |
Hb_156947_010 |
0.0834463742 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 9 |
Hb_143766_050 |
0.0841606996 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 10 |
Hb_005511_160 |
0.0849575842 |
- |
- |
OB-fold nucleic acid binding domain-containing protein [Theobroma cacao] |
| 11 |
Hb_000580_080 |
0.085430738 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 12 |
Hb_002375_090 |
0.0869208485 |
- |
- |
- |
| 13 |
Hb_002259_070 |
0.088093904 |
- |
- |
taz protein, putative [Ricinus communis] |
| 14 |
Hb_003203_040 |
0.0896240975 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 15 |
Hb_002660_030 |
0.0900716666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000136_170 |
0.0904394861 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 17 |
Hb_002701_240 |
0.0909201547 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 18 |
Hb_005618_140 |
0.0921535435 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 19 |
Hb_012753_160 |
0.0928132388 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 20 |
Hb_000526_040 |
0.0932164194 |
- |
- |
60S ribosomal protein L6, putative [Ricinus communis] |