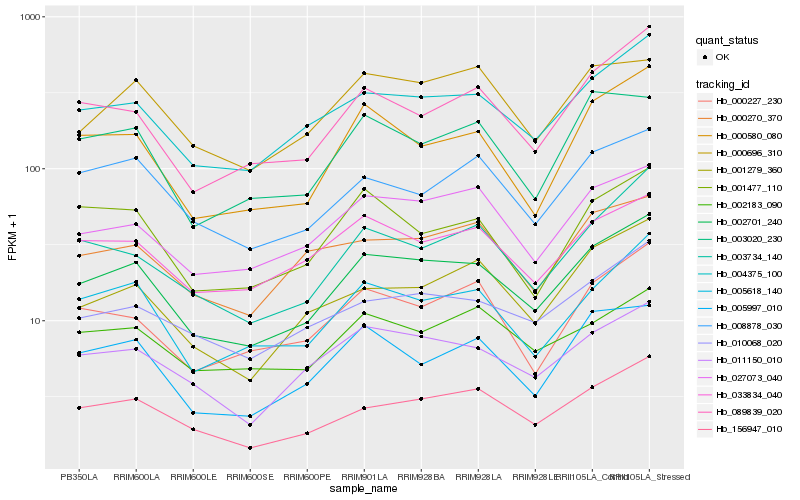
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_240 |
0.0 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 2 |
Hb_004375_100 |
0.0616508855 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 3 |
Hb_156947_010 |
0.0699078113 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 4 |
Hb_011150_010 |
0.0741211546 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 5 |
Hb_005618_140 |
0.0771714328 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 6 |
Hb_027073_040 |
0.0781388804 |
- |
- |
TGF-beta-inducible nuclear protein, putative [Ricinus communis] |
| 7 |
Hb_010068_020 |
0.084117333 |
- |
- |
PREDICTED: uncharacterized protein LOC105632966 [Jatropha curcas] |
| 8 |
Hb_033834_040 |
0.087988581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005997_010 |
0.0892491839 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 10 |
Hb_000580_080 |
0.0895246205 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 11 |
Hb_003734_140 |
0.0909201547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_089839_020 |
0.0923423626 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 13 |
Hb_001279_360 |
0.0931889613 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000227_230 |
0.0943424417 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 15 |
Hb_008878_030 |
0.0948710453 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Musa acuminata subsp. malaccensis] |
| 16 |
Hb_000270_370 |
0.0959596777 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 17 |
Hb_000696_310 |
0.0959937048 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 18 |
Hb_003020_230 |
0.0966342453 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 19 |
Hb_002183_090 |
0.0969486149 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 20 |
Hb_001477_110 |
0.0970863482 |
- |
- |
prohibitin, putative [Ricinus communis] |