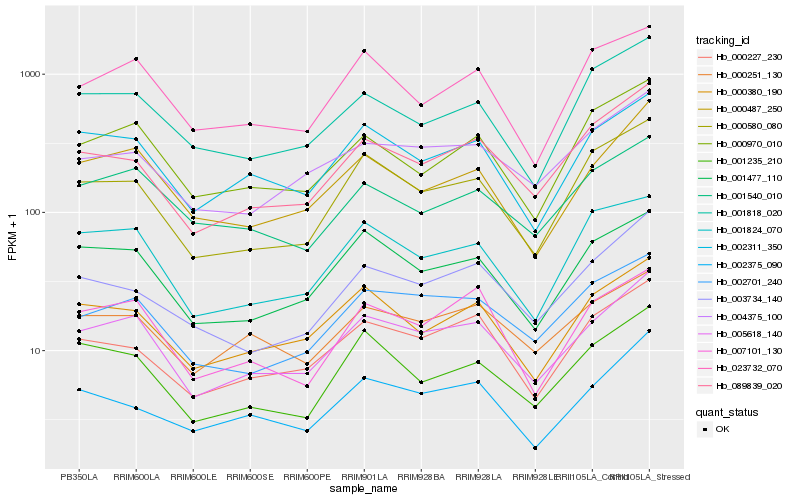
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005618_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 2 |
Hb_002311_350 |
0.074958232 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 3 |
Hb_000487_250 |
0.0759710645 |
- |
- |
60S ribosomal protein L27e, putative [Ricinus communis] |
| 4 |
Hb_002701_240 |
0.0771714328 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 5 |
Hb_001477_110 |
0.0788809182 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 6 |
Hb_000380_190 |
0.0801214351 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 7 |
Hb_089839_020 |
0.0820743326 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 8 |
Hb_000227_230 |
0.0821815067 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 9 |
Hb_004375_100 |
0.0825452454 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 10 |
Hb_000580_080 |
0.0836376474 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 11 |
Hb_000251_130 |
0.0849170375 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 12 |
Hb_007101_130 |
0.0894017371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000970_010 |
0.0899826968 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 14 |
Hb_001540_010 |
0.0919558324 |
- |
- |
PREDICTED: uncharacterized protein LOC105639920 [Jatropha curcas] |
| 15 |
Hb_003734_140 |
0.0921535435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001824_070 |
0.0924580124 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 17 |
Hb_001235_210 |
0.0936668824 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_023732_070 |
0.0945724905 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 19 |
Hb_002375_090 |
0.096358856 |
- |
- |
- |
| 20 |
Hb_001818_020 |
0.0964542764 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |