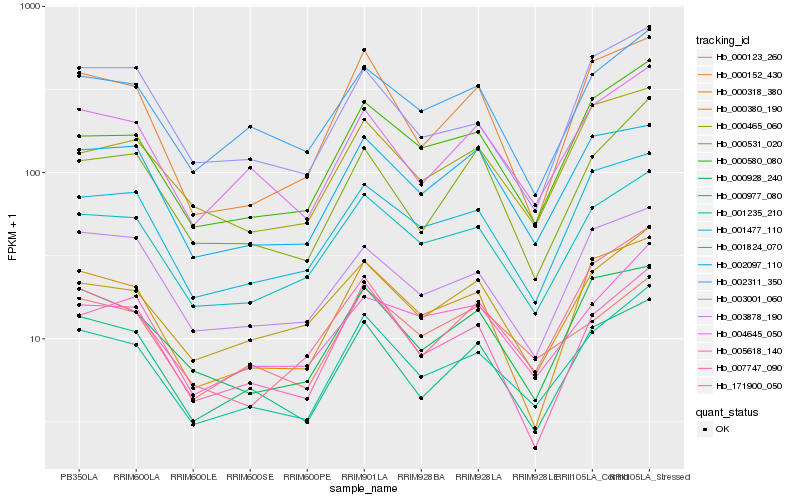
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_210 |
0.0 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_001477_110 |
0.0660783977 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 3 |
Hb_004645_050 |
0.0670117295 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 4 |
Hb_000580_080 |
0.0737088461 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 5 |
Hb_002311_350 |
0.0781186343 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 6 |
Hb_001824_070 |
0.0815074492 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 7 |
Hb_000380_190 |
0.082104221 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 8 |
Hb_007747_090 |
0.0849425121 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 9 |
Hb_000977_080 |
0.0863366593 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_000928_240 |
0.0867585317 |
- |
- |
PREDICTED: uncharacterized protein LOC105632515 [Jatropha curcas] |
| 11 |
Hb_000318_380 |
0.0907154509 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 3-like [Jatropha curcas] |
| 12 |
Hb_000465_060 |
0.0918598126 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000152_430 |
0.0919585801 |
- |
- |
PREDICTED: 40S ribosomal protein S20-1 [Jatropha curcas] |
| 14 |
Hb_000123_260 |
0.0933462573 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 15 |
Hb_000531_020 |
0.0933558306 |
- |
- |
PREDICTED: uncharacterized protein At4g28440-like [Populus euphratica] |
| 16 |
Hb_005618_140 |
0.0936668824 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 17 |
Hb_003878_190 |
0.0942080857 |
- |
- |
hypothetical protein POPTR_0011s11040g [Populus trichocarpa] |
| 18 |
Hb_003001_060 |
0.0942581837 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 19 |
Hb_002097_110 |
0.0946415528 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_171900_050 |
0.0953146018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |