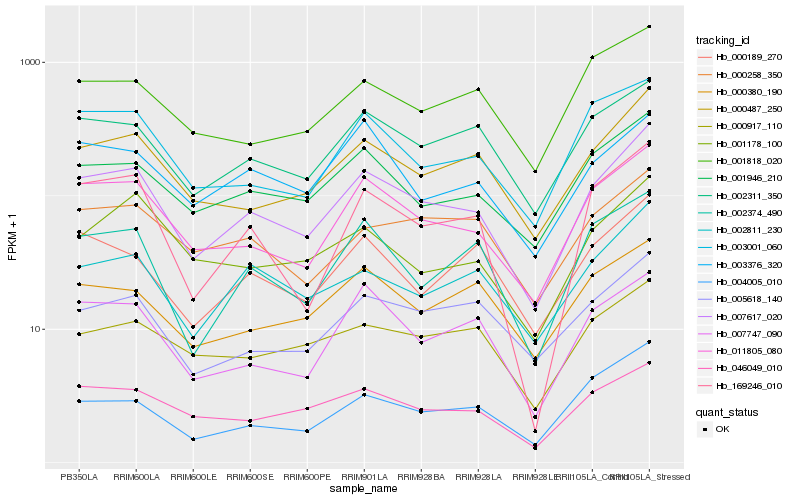
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007617_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011805_080 |
0.0735856738 |
- |
- |
PREDICTED: uncharacterized protein LOC100242357 [Vitis vinifera] |
| 3 |
Hb_000487_250 |
0.088176521 |
- |
- |
60S ribosomal protein L27e, putative [Ricinus communis] |
| 4 |
Hb_001946_210 |
0.0950389632 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |
| 5 |
Hb_002311_350 |
0.100421713 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 6 |
Hb_002811_230 |
0.1064180632 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 7 |
Hb_169246_010 |
0.1103525831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003001_060 |
0.1112555148 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 9 |
Hb_005618_140 |
0.11142769 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 10 |
Hb_046049_010 |
0.1118787374 |
- |
- |
PREDICTED: far upstream element-binding protein 2 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002374_490 |
0.1127176341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007747_090 |
0.1131128739 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 13 |
Hb_004005_010 |
0.1154957731 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 14 |
Hb_000189_270 |
0.1174013064 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 15 |
Hb_001178_100 |
0.1200606169 |
- |
- |
60S acidic ribosomal protein P2-4 [Theobroma cacao] |
| 16 |
Hb_000258_350 |
0.1205520844 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 17 |
Hb_001818_020 |
0.1218890497 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1 [Phoenix dactylifera] |
| 18 |
Hb_000917_110 |
0.1240054545 |
- |
- |
PREDICTED: nudix hydrolase 1 [Jatropha curcas] |
| 19 |
Hb_000380_190 |
0.1243434865 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 20 |
Hb_003376_320 |
0.1243634736 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |