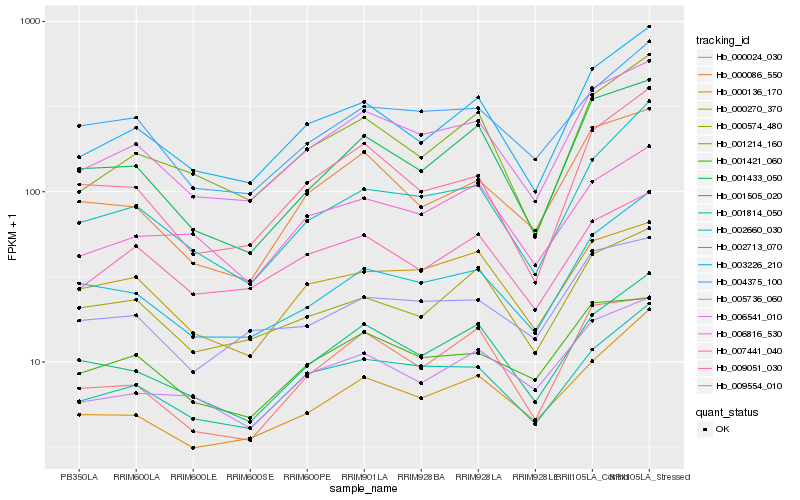
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_530 |
0.0 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 2 |
Hb_001814_050 |
0.0567212563 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_001505_020 |
0.0614329588 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 9 [Jatropha curcas] |
| 4 |
Hb_003226_210 |
0.0657438187 |
- |
- |
hypothetical protein PRUPE_ppa013414mg [Prunus persica] |
| 5 |
Hb_001214_160 |
0.0697416815 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 6 |
Hb_002713_070 |
0.0714392964 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 7 |
Hb_001421_060 |
0.0726099406 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |
| 8 |
Hb_000086_550 |
0.074299751 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 9 |
Hb_001433_050 |
0.0745779987 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 10 |
Hb_000024_030 |
0.0746130836 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 5-like [Populus euphratica] |
| 11 |
Hb_004375_100 |
0.0757657115 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 12 |
Hb_006541_010 |
0.0777224996 |
- |
- |
PREDICTED: uncharacterized protein LOC105637261 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000136_170 |
0.0780633881 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 14 |
Hb_009051_030 |
0.0790815872 |
- |
- |
PREDICTED: thioredoxin O1, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_005736_060 |
0.0795667554 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 16 |
Hb_000574_480 |
0.0800235674 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 17 |
Hb_009554_010 |
0.0807169089 |
- |
- |
PREDICTED: multifunctional methyltransferase subunit TRM112-like protein At1g22270 [Jatropha curcas] |
| 18 |
Hb_000270_370 |
0.0811185384 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 19 |
Hb_007441_040 |
0.0812989628 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 20 |
Hb_002660_030 |
0.0813845443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |