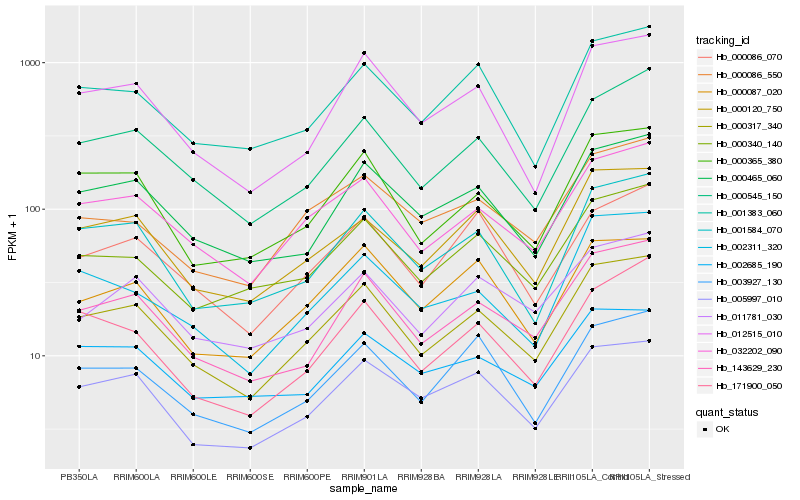
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_032202_090 |
0.044973323 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Jatropha curcas] |
| 3 |
Hb_143629_230 |
0.0572230808 |
- |
- |
ring finger, putative [Ricinus communis] |
| 4 |
Hb_000365_380 |
0.062672146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000545_150 |
0.0655181189 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 6 |
Hb_000465_060 |
0.0664557588 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000120_750 |
0.067177466 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_001584_070 |
0.0688649813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005997_010 |
0.0709049044 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 10 |
Hb_012515_010 |
0.0750697935 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |
| 11 |
Hb_001383_060 |
0.0769899873 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 12 |
Hb_000340_140 |
0.0789543372 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Vitis vinifera] |
| 13 |
Hb_003927_130 |
0.0818775407 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 14 |
Hb_011781_030 |
0.0838627885 |
- |
- |
Frataxin, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_000086_550 |
0.084229975 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 16 |
Hb_002311_320 |
0.0843692665 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 20a [Jatropha curcas] |
| 17 |
Hb_171900_050 |
0.0848513561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000087_020 |
0.0850732285 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase 2, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002685_190 |
0.0855391527 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000086_070 |
0.0858763271 |
- |
- |
Peptidase S24/S26A/S26B/S26C family protein isoform 1 [Theobroma cacao] |