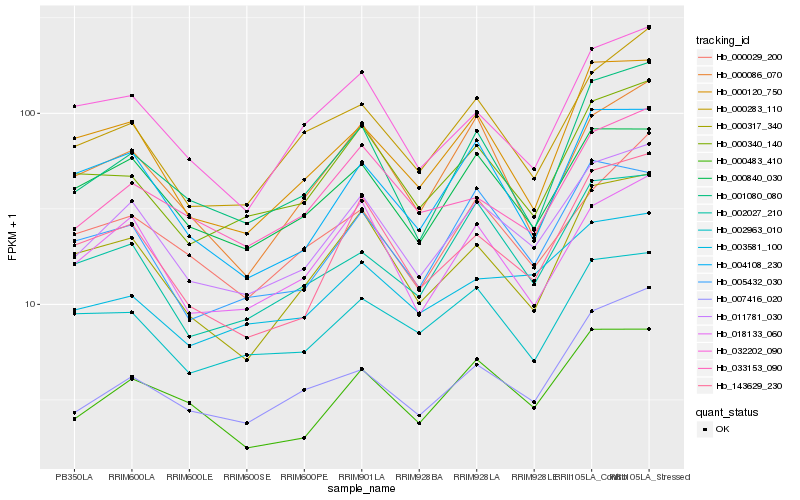
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011781_030 |
0.0 |
- |
- |
Frataxin, mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_000483_410 |
0.0762151723 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_018133_060 |
0.0787094177 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit RPA12-like [Jatropha curcas] |
| 4 |
Hb_143629_230 |
0.079859335 |
- |
- |
ring finger, putative [Ricinus communis] |
| 5 |
Hb_000840_030 |
0.081271808 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 6 |
Hb_000340_140 |
0.081299928 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Vitis vinifera] |
| 7 |
Hb_000317_340 |
0.0838627885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001080_080 |
0.0841221576 |
- |
- |
small nuclear ribonucleoprotein, putative [Ricinus communis] |
| 9 |
Hb_005432_030 |
0.0875477894 |
- |
- |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Gossypium raimondii] |
| 10 |
Hb_000120_750 |
0.0878999028 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000283_110 |
0.0883770473 |
- |
- |
PREDICTED: probable prefoldin subunit 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_033153_090 |
0.0899837401 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 13 |
Hb_000029_200 |
0.0910755957 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 14 |
Hb_002027_210 |
0.091232647 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 15 |
Hb_032202_090 |
0.0920215631 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Jatropha curcas] |
| 16 |
Hb_004108_230 |
0.0921064568 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 17 |
Hb_007416_020 |
0.0926129506 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Cucumis sativus] |
| 18 |
Hb_003581_100 |
0.093324854 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000086_070 |
0.0935007041 |
- |
- |
Peptidase S24/S26A/S26B/S26C family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_002963_010 |
0.0937843791 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |