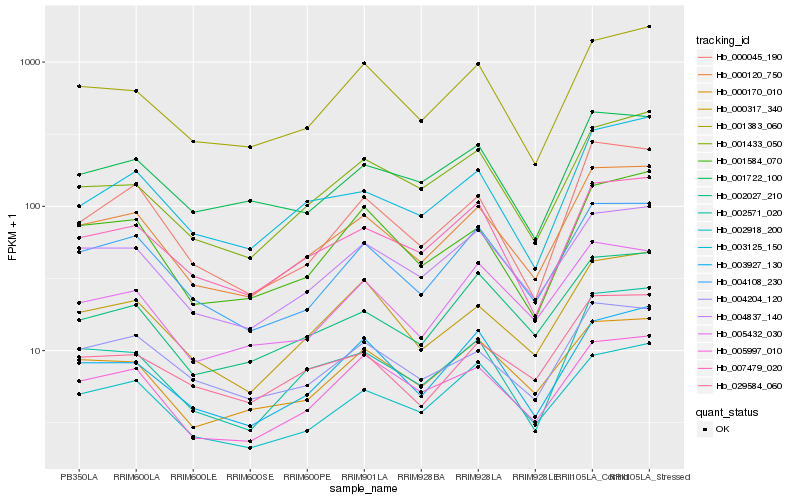
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_750 |
0.0 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_004108_230 |
0.056948668 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 3 |
Hb_004837_140 |
0.0638288819 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 4 |
Hb_007479_020 |
0.0660785733 |
- |
- |
PREDICTED: CMP-sialic acid transporter 5 [Jatropha curcas] |
| 5 |
Hb_001383_060 |
0.0667920987 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 6 |
Hb_000317_340 |
0.067177466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004204_120 |
0.0722164493 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 8 |
Hb_002918_200 |
0.0722703429 |
- |
- |
- |
| 9 |
Hb_000170_010 |
0.0726685046 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001584_070 |
0.0731448997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002027_210 |
0.0745790519 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 12 |
Hb_003927_130 |
0.0746559881 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 13 |
Hb_029584_060 |
0.0747187857 |
- |
- |
Grave disease carrier protein, putative [Ricinus communis] |
| 14 |
Hb_000045_190 |
0.0750143469 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit F [Jatropha curcas] |
| 15 |
Hb_003125_150 |
0.0752459145 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma [Jatropha curcas] |
| 16 |
Hb_001433_050 |
0.0757720417 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 17 |
Hb_002571_020 |
0.0764142812 |
- |
- |
PREDICTED: uncharacterized protein LOC105132457 isoform X12 [Populus euphratica] |
| 18 |
Hb_005997_010 |
0.0798348147 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 19 |
Hb_001722_100 |
0.0803918784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005432_030 |
0.0806174219 |
- |
- |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Gossypium raimondii] |