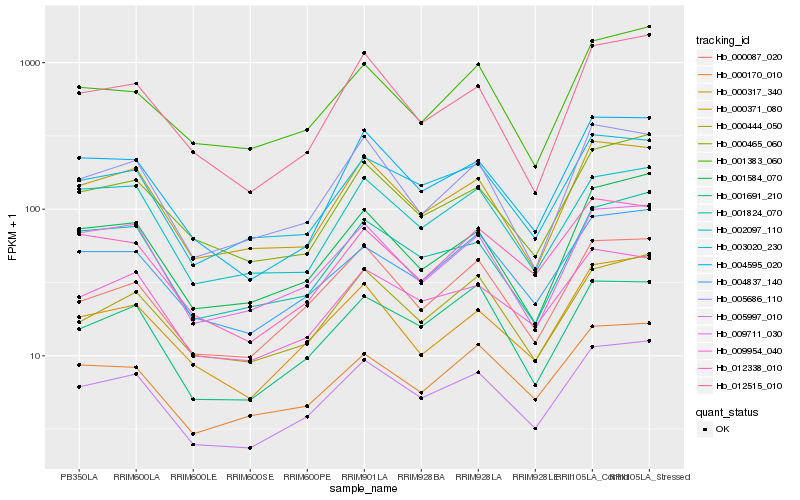
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005997_010 |
0.0 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 2 |
Hb_003020_230 |
0.0536654709 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 3 |
Hb_004837_140 |
0.0593614734 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 4 |
Hb_009954_040 |
0.0618872515 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 5 |
Hb_000087_020 |
0.0637571128 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001824_070 |
0.0648597847 |
- |
- |
H/ACA ribonucleoprotein complex subunit, putative [Ricinus communis] |
| 7 |
Hb_000371_080 |
0.0650686703 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 8 |
Hb_000465_060 |
0.0650829694 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000444_050 |
0.0665101448 |
transcription factor |
TF Family: HMG |
PREDICTED: uncharacterized protein LOC105648212 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000170_010 |
0.0682998659 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001691_210 |
0.0700928162 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 12 |
Hb_000317_340 |
0.0709049044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004595_020 |
0.0722095225 |
- |
- |
PREDICTED: probable prefoldin subunit 5 [Jatropha curcas] |
| 14 |
Hb_001584_070 |
0.0728146409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009711_030 |
0.0732137891 |
- |
- |
PREDICTED: uncharacterized protein LOC105647792 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005686_110 |
0.0759745408 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 17 |
Hb_002097_110 |
0.0782505172 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_012515_010 |
0.0784171592 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |
| 19 |
Hb_001383_060 |
0.0787910021 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 20 |
Hb_012338_010 |
0.0789215548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |