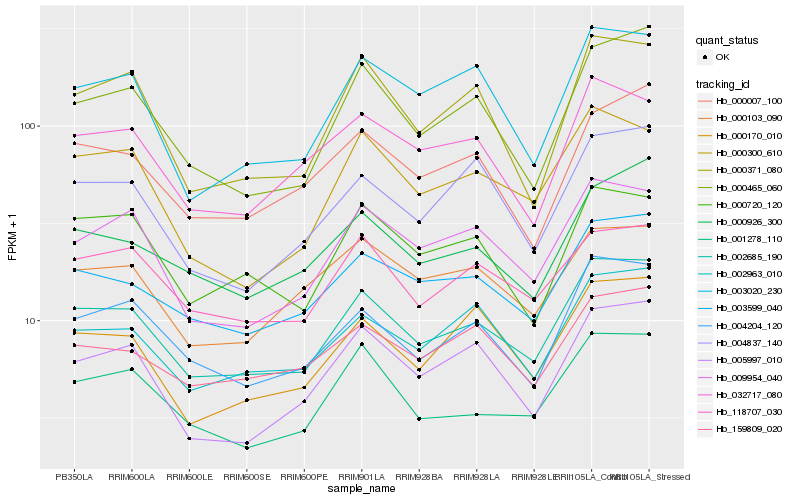
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002685_190 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003599_040 |
0.0577659404 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_004204_120 |
0.0601555262 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 4 |
Hb_002963_010 |
0.0686180389 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000465_060 |
0.068825817 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_009954_040 |
0.0699467478 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 7 |
Hb_001278_110 |
0.0717247866 |
- |
- |
PREDICTED: uncharacterized protein LOC104613296 isoform X3 [Nelumbo nucifera] |
| 8 |
Hb_000926_300 |
0.0740547193 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 9 |
Hb_118707_030 |
0.0754129317 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 19a-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000007_100 |
0.0770712867 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 11 |
Hb_003020_230 |
0.0773075976 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 12 |
Hb_000300_610 |
0.0781113414 |
- |
- |
cytosolic copper/zinc superoxide dismutase [Hevea brasiliensis] |
| 13 |
Hb_159809_020 |
0.0788030473 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000720_120 |
0.0795604243 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 15 |
Hb_000170_010 |
0.0795912871 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_005997_010 |
0.0803194804 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 17 |
Hb_004837_140 |
0.0804989868 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 18 |
Hb_032717_080 |
0.0806109525 |
- |
- |
gamma-soluble nsf attachment protein, putative [Ricinus communis] |
| 19 |
Hb_000371_080 |
0.0818765947 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 20 |
Hb_000103_090 |
0.0818900963 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |