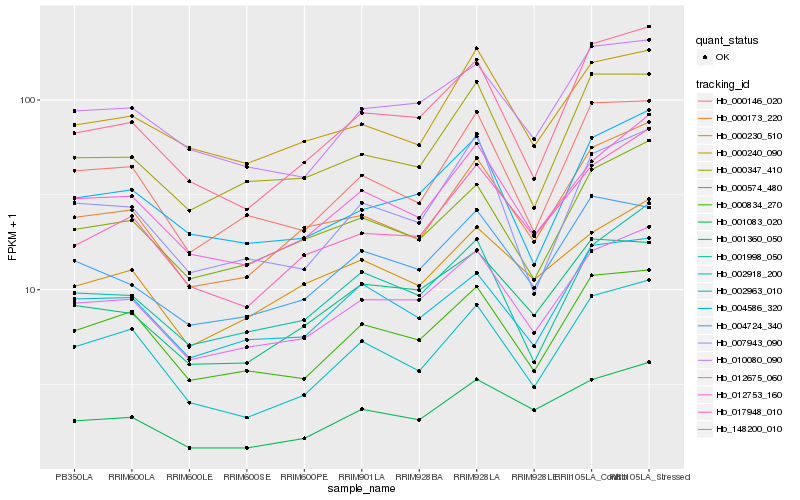
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012675_060 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 2 |
Hb_012753_160 |
0.0495980979 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 3 |
Hb_010080_090 |
0.0546812144 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 [Jatropha curcas] |
| 4 |
Hb_000834_270 |
0.0562905167 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004586_320 |
0.0569662945 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 6 |
Hb_000574_480 |
0.060464757 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 7 |
Hb_000173_220 |
0.0650501683 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 8 |
Hb_001998_050 |
0.067935002 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_002963_010 |
0.0687754963 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_148200_010 |
0.0688519199 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 11 |
Hb_001083_020 |
0.0694611447 |
- |
- |
- |
| 12 |
Hb_000146_020 |
0.0696815367 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001360_050 |
0.0701897063 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000230_510 |
0.0706046699 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 15 |
Hb_000347_410 |
0.0723726146 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 16 |
Hb_004724_340 |
0.0727389571 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 17 |
Hb_017948_010 |
0.0727876661 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 18 |
Hb_000240_090 |
0.0763662857 |
- |
- |
Rab5 [Hevea brasiliensis] |
| 19 |
Hb_002918_200 |
0.0767338827 |
- |
- |
- |
| 20 |
Hb_007943_090 |
0.0768760244 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |