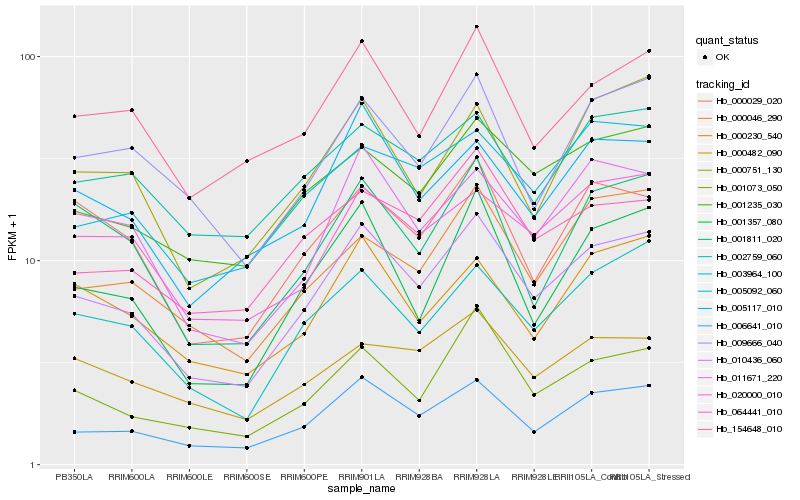
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010436_060 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0003801mg, partial [Citrus sinensis] |
| 2 |
Hb_006641_010 |
0.0584006542 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 3 |
Hb_003964_100 |
0.0872121319 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 4 |
Hb_011671_220 |
0.0883658334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001235_030 |
0.0884309633 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 6 |
Hb_001073_050 |
0.0886897732 |
- |
- |
- |
| 7 |
Hb_005117_010 |
0.0918497105 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_001357_080 |
0.0918864367 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 9 |
Hb_154648_010 |
0.0952794755 |
- |
- |
PREDICTED: uncharacterized protein LOC105634412 isoform X2 [Jatropha curcas] |
| 10 |
Hb_020000_010 |
0.0957348746 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 11 |
Hb_064441_010 |
0.09590373 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005092_060 |
0.1004798955 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 13 |
Hb_009666_040 |
0.1008300825 |
- |
- |
glutathione transferase, putative [Ricinus communis] |
| 14 |
Hb_002759_060 |
0.1014511393 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 15 |
Hb_000046_290 |
0.1017471545 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 16 |
Hb_000029_020 |
0.1018264808 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 17 |
Hb_000230_540 |
0.1034940377 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 18 |
Hb_001811_020 |
0.1038772511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000482_090 |
0.1041653027 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 20 |
Hb_000751_130 |
0.1054364595 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |