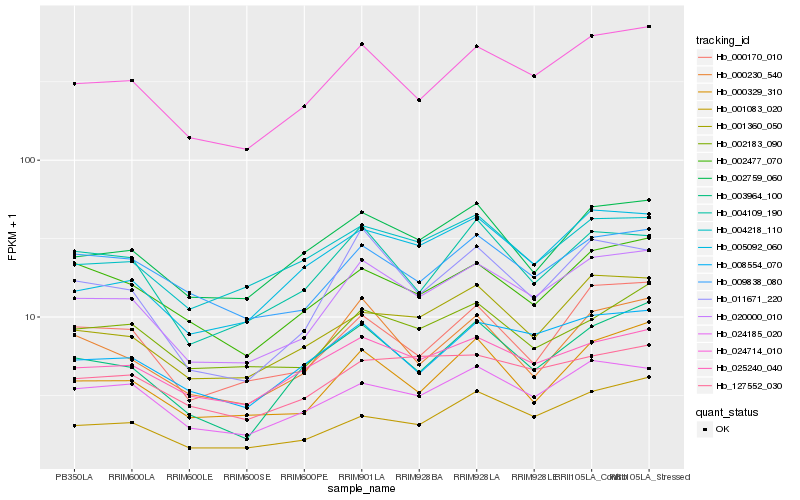
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020000_010 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 2 |
Hb_024714_010 |
0.0490850305 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_024185_020 |
0.078671223 |
- |
- |
- |
| 4 |
Hb_008554_070 |
0.0794027373 |
- |
- |
PREDICTED: peroxisome biogenesis protein 3-2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001360_050 |
0.0806866805 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011671_220 |
0.0814764209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002759_060 |
0.0833173082 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 8 |
Hb_025240_040 |
0.0847385175 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |
| 9 |
Hb_001083_020 |
0.0876051816 |
- |
- |
- |
| 10 |
Hb_000230_540 |
0.0890477381 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 11 |
Hb_000329_310 |
0.0896020747 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 12 |
Hb_009838_080 |
0.0913735086 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_004109_190 |
0.0918157412 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 25-like isoform X2 [Populus euphratica] |
| 14 |
Hb_005092_060 |
0.0922666316 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 15 |
Hb_003964_100 |
0.0924091371 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 16 |
Hb_000170_010 |
0.0924914983 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_127552_030 |
0.0932401807 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 18 |
Hb_004218_110 |
0.0934008978 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 19 |
Hb_002183_090 |
0.0942772495 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 20 |
Hb_002477_070 |
0.0948391445 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |