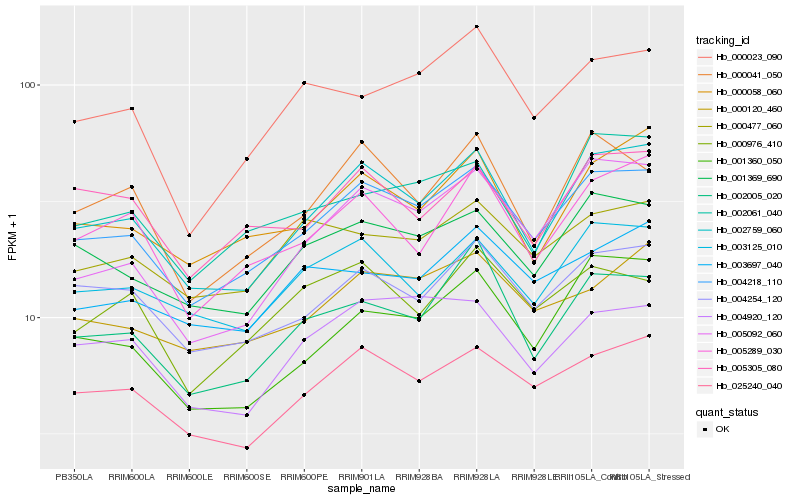
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_110 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 2 |
Hb_002759_060 |
0.0484363584 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 3 |
Hb_025240_040 |
0.0542122899 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |
| 4 |
Hb_001369_690 |
0.0592381379 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 5 |
Hb_004254_120 |
0.0596449915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000120_460 |
0.0700452713 |
- |
- |
PREDICTED: thioredoxin reductase NTRB-like [Jatropha curcas] |
| 7 |
Hb_002005_020 |
0.071636016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003125_010 |
0.0727099102 |
- |
- |
PREDICTED: uncharacterized protein LOC105640205 [Jatropha curcas] |
| 9 |
Hb_005092_060 |
0.0750672254 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 10 |
Hb_000058_060 |
0.0756133371 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase IBR5 [Jatropha curcas] |
| 11 |
Hb_005289_030 |
0.0758124503 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_000041_050 |
0.0771331385 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 13 |
Hb_002061_040 |
0.0777660815 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 14 |
Hb_000976_410 |
0.078740478 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 15 |
Hb_000477_060 |
0.0790760617 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 16 |
Hb_000023_090 |
0.0794955818 |
- |
- |
mak, putative [Ricinus communis] |
| 17 |
Hb_004920_120 |
0.0797640608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003697_040 |
0.0799301671 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 19 |
Hb_001360_050 |
0.0806787581 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005305_080 |
0.0809896944 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |