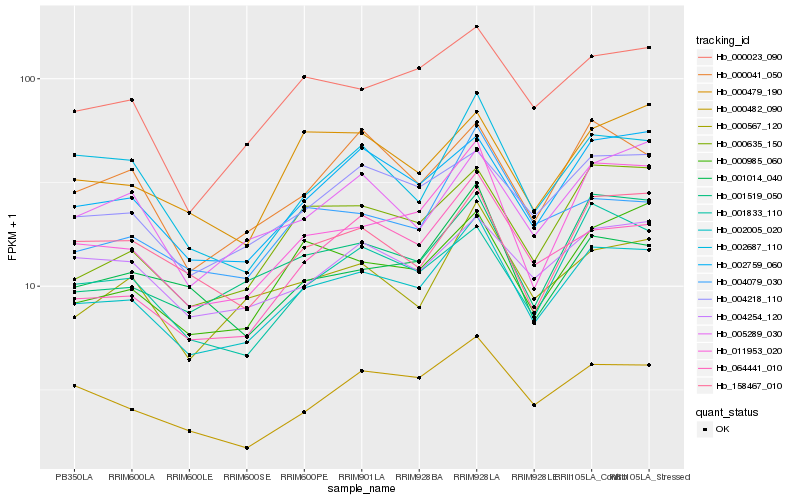
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004218_110 |
0.071636016 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 3 |
Hb_000023_090 |
0.0717260459 |
- |
- |
mak, putative [Ricinus communis] |
| 4 |
Hb_002759_060 |
0.0718507204 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 5 |
Hb_011953_020 |
0.0738322595 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000567_120 |
0.0782914368 |
- |
- |
PREDICTED: transcription factor IIIB 50 kDa subunit-like [Jatropha curcas] |
| 7 |
Hb_001519_050 |
0.0785457773 |
- |
- |
PREDICTED: protein RMD5 homolog A-like [Jatropha curcas] |
| 8 |
Hb_000482_090 |
0.0801962498 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 9 |
Hb_000635_150 |
0.081287369 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 10 |
Hb_000985_060 |
0.0828441369 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004254_120 |
0.0835743503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001014_040 |
0.0855503191 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 13 |
Hb_005289_030 |
0.0861089923 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_158467_010 |
0.0865790337 |
- |
- |
transporter, putative [Ricinus communis] |
| 15 |
Hb_001833_110 |
0.0884323918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002687_110 |
0.0889767981 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C343.04c isoform X1 [Jatropha curcas] |
| 17 |
Hb_064441_010 |
0.0889780077 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000041_050 |
0.0889916643 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 19 |
Hb_000479_190 |
0.0892506379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004079_030 |
0.0895171389 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |