| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
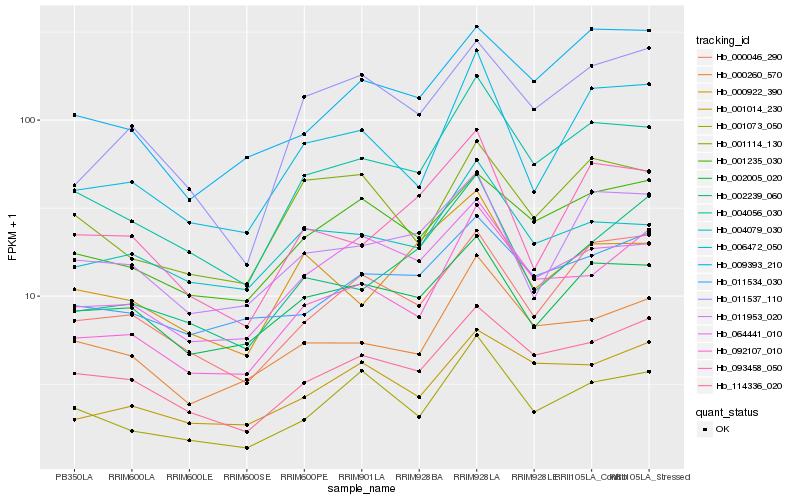
Hb_004056_030 |
0.0 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 2 |
Hb_114336_020 |
0.0941106729 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 3 |
Hb_092107_010 |
0.0968170355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001073_050 |
0.0975957456 |
- |
- |
- |
| 5 |
Hb_000260_570 |
0.0989491294 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_064441_010 |
0.1015962078 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_006472_050 |
0.1087422174 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 8 |
Hb_001235_030 |
0.1109049341 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 9 |
Hb_000046_290 |
0.1123199866 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 10 |
Hb_009393_210 |
0.1176716142 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 11 |
Hb_093458_050 |
0.1179253662 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 12 |
Hb_011534_030 |
0.1198565342 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 13 |
Hb_001114_130 |
0.1208164042 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001014_230 |
0.1212330717 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 15 |
Hb_004079_030 |
0.1226614242 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 16 |
Hb_011953_020 |
0.1228027052 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 17 |
Hb_000922_390 |
0.1229293553 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 18 |
Hb_011537_110 |
0.1238997069 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 19 |
Hb_002005_020 |
0.1249407071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002239_060 |
0.1252039493 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |