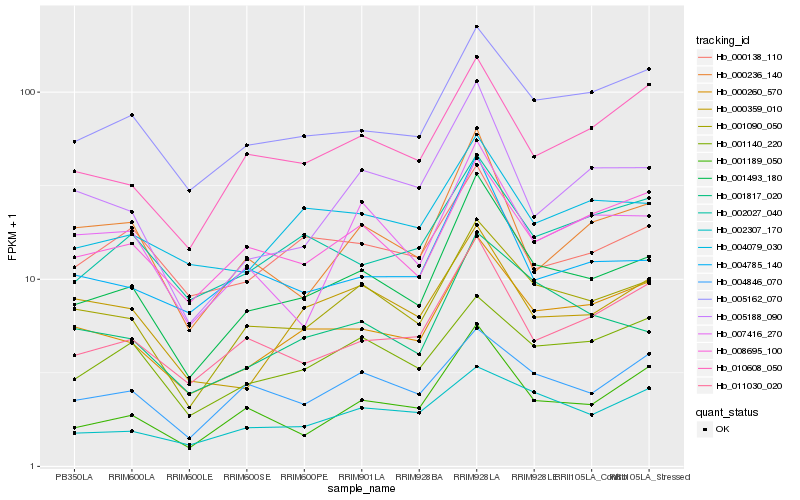
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001493_180 |
0.0 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 2 |
Hb_001090_050 |
0.0709723281 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000260_570 |
0.0897819275 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_005162_070 |
0.0986218841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001817_020 |
0.098763097 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 6 |
Hb_001189_050 |
0.1062027226 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 7 |
Hb_011030_020 |
0.1063835295 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 8 |
Hb_004785_140 |
0.1082057592 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_005188_090 |
0.1143602825 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 10 |
Hb_004846_070 |
0.1156437477 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 11 |
Hb_000236_140 |
0.116332013 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 12 |
Hb_008695_100 |
0.1176054891 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004079_030 |
0.1197493615 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 14 |
Hb_010608_050 |
0.1212861371 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 15 |
Hb_007416_270 |
0.122010526 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 16 |
Hb_002027_040 |
0.122796282 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 17 |
Hb_000359_010 |
0.1243803188 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 18 |
Hb_001140_220 |
0.1245942704 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_002307_170 |
0.1261359454 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_000138_110 |
0.1287580368 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |