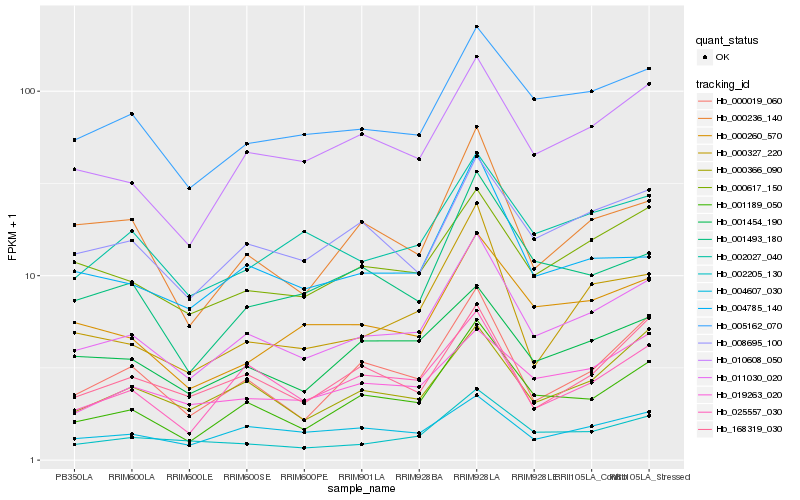
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011030_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 2 |
Hb_005162_070 |
0.0780667562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001189_050 |
0.0793248264 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 4 |
Hb_010608_050 |
0.0803097635 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 5 |
Hb_002205_130 |
0.0869291643 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 6 |
Hb_008695_100 |
0.0897883011 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000617_150 |
0.0901886429 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 8 |
Hb_004607_030 |
0.0931553365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001454_190 |
0.0941252591 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 10 |
Hb_000019_060 |
0.0953374994 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 11 |
Hb_025557_030 |
0.0987702386 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002027_040 |
0.0987938787 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 13 |
Hb_000366_090 |
0.0999943855 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000260_570 |
0.1018048308 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_004785_140 |
0.1022956336 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000327_220 |
0.1040293681 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 17 |
Hb_000236_140 |
0.1040734237 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 18 |
Hb_019263_020 |
0.1057114183 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001493_180 |
0.1063835295 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 20 |
Hb_168319_030 |
0.1066978724 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |